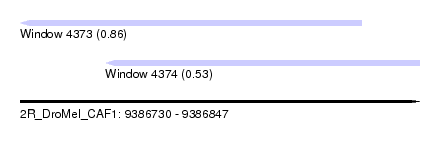
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,386,730 – 9,386,847 |
| Length | 117 |
| Max. P | 0.862024 |

| Location | 9,386,730 – 9,386,830 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.80 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -23.63 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
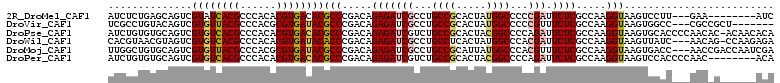
>2R_DroMel_CAF1 9386730 100 - 20766785 AUCUCUGAGCAGUCGUAUCACGCCCACACGUGACACGCCCGACAGAGAUCGCCUGCCGCACUAUGGCCCCCGAUUCUCGCCAAGGUAAGUCCUU---GAA--------AUC .((((((..(.(.(((.(((((......))))).))).).).))))))(((...((((.....))))...))).......(((((.....))))---)..--------... ( -28.30) >DroVir_CAF1 10152 101 - 1 UCGCCUGUACAGUCGUGUUACGCCCACGCGUGAUACGCCCGACAGAGAUCGCCUGCCGCACUAUGGCCCCCGUUUCUCGCCAAGGUAAGUGGCC---CGCCGCU------- ..((((((...(.((((((((((....)))))))))))...)))).....((..((((((((.((((...........)))).)))..))))).---.)).)).------- ( -35.80) >DroPse_CAF1 8626 110 - 1 AUCUGUGUGCAGUCGUGUCACGCCCACACGUGACACGCCCGACAGAGAUCGUCUGCCGCACUACGGCCCCAGAUUCUCGCCAAGGUAAGUGCACCCCAACAC-ACAACACA ...((((((.....((((((((......))))))))(((.....((((((....((((.....))))....)).)))).....))).............)))-)))..... ( -35.60) >DroWil_CAF1 11119 107 - 1 CACGUAACGUAGUCGUGUCACGCCCACACGUGAUACACCCGACAGAGAUCGCCUGCCUCACUAUGGCCCACGAUUCUCGCCAAGGUAAGUUAUC---AACAG-CCAAGAGA ...(((((......((((((((......))))))))(((.....(((((((...(((.......)))...))).)))).....)))..))))).---.....-........ ( -28.80) >DroMoj_CAF1 19339 108 - 1 UUGGCUGUGCAGUCGUGUUACGCCCACGCGUGAUACGCCCGACAGAGAUCGCCUGCCGCAUUAUGGCCCACGUUUCUCGCCAAGGUAAGUGACC---AACCGACCAAUCGA ((((((((.(.(.((((((((((....)))))))))).).))))((((.((...((((.....))))...))..)))))))))(((..((....---.))..)))...... ( -36.30) >DroPer_CAF1 10411 103 - 1 AUCUGUGUGCAGUCGUGUCACGCCCACACGUGACACGCCCGACAGAGAUCGUCUGCCGCACUACGGCCCCAGAUUCUCGCCAAGGUAAGUCCACCCCAAC--------ACA ....((((...(.(((((((((......))))))))))..(((.((((((....((((.....))))....)).))))((....))..))).......))--------)). ( -33.60) >consensus AUCUCUGUGCAGUCGUGUCACGCCCACACGUGACACGCCCGACAGAGAUCGCCUGCCGCACUAUGGCCCCCGAUUCUCGCCAAGGUAAGUGCAC___AAC_G______ACA ..............((((((((......))))))))(((.....(((((((...((((.....))))...))).)))).....)))......................... (-23.63 = -23.93 + 0.31)
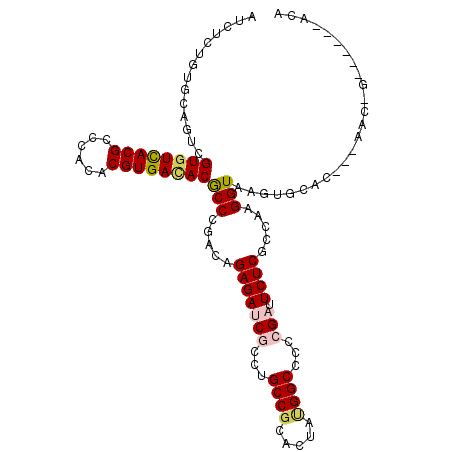
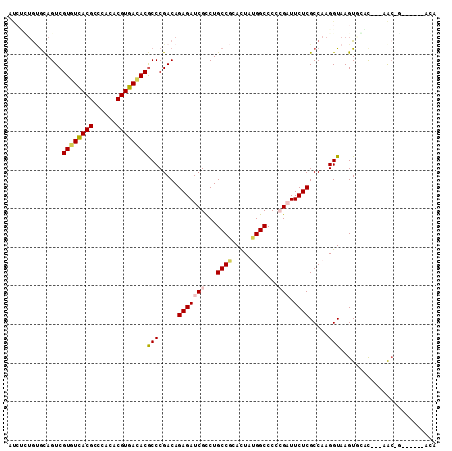
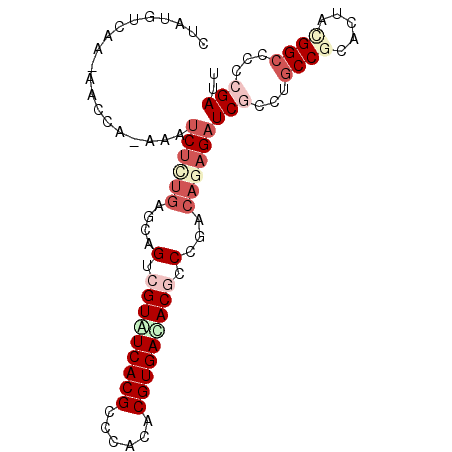
| Location | 9,386,755 – 9,386,847 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 84.34 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -17.39 |
| Energy contribution | -19.50 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9386755 92 - 20766785 CUAUGUCAAAAACCA-AAAUCUCUGAGCAGUCGUAUCACGCCCACACGUGACACGCCCGACAGAGAUCGCCUGCCGCACUAUGGCCCCCGAUU ...............-...((((((..(.(.(((.(((((......))))).))).).).))))))(((...((((.....))))...))).. ( -22.60) >DroPse_CAF1 8661 92 - 1 CAAUGUACA-AACCAAAUAUCUGUGUGCAGUCGUGUCACGCCCACACGUGACACGCCCGACAGAGAUCGUCUGCCGCACUACGGCCCCAGAUU .........-........((((.(((.(.(.(((((((((......))))))))).).)))).)))).((((((((.....))))...)))). ( -28.30) >DroSim_CAF1 8952 91 - 1 CUAUGUCAA-AACCA-AAAUCUCUGAGCAGUCGUAUCACGCCCACACGUGAUACUCCCGACAGAGAUCGCCUGCCGCACUAUGGCCCCCGAUU .........-.....-...((((((..(.(..((((((((......))))))))..).).))))))(((...((((.....))))...))).. ( -24.50) >DroWil_CAF1 11151 83 - 1 -------AA-AA-AA-AACACGUAACGUAGUCGUGUCACGCCCACACGUGAUACACCCGACAGAGAUCGCCUGCCUCACUAUGGCCCACGAUU -------..-..-..-....(((..((((((.((((((((......))))))))........(((.........)))))))))....)))... ( -20.20) >DroYak_CAF1 8999 91 - 1 CUAUGUCAA-AUGCA-AAAUCUCUGAGCAGUCGUAUCACGCCCACACGUGACACGCCCGACAGAGAUCGCCUGCCUCACUACGGGCCCCGAUU ....(((..-.....-..(((((((..(.(.(((.(((((......))))).))).).).))))))).(((((........)))))...))). ( -25.20) >DroPer_CAF1 10439 92 - 1 CAAUGUACA-AACCAAAUAUCUGUGUGCAGUCGUGUCACGCCCACACGUGACACGCCCGACAGAGAUCGUCUGCCGCACUACGGCCCCAGAUU .........-........((((.(((.(.(.(((((((((......))))))))).).)))).)))).((((((((.....))))...)))). ( -28.30) >consensus CUAUGUCAA_AACCA_AAAUCUCUGAGCAGUCGUAUCACGCCCACACGUGACACGCCCGACAGAGAUCGCCUGCCGCACUACGGCCCCCGAUU ...................((((((....(.(((((((((......))))))))).)...))))))(((...((((.....))))...))).. (-17.39 = -19.50 + 2.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:35 2006