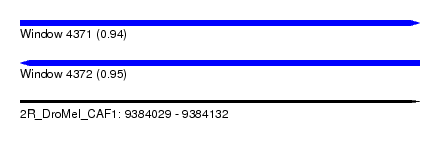
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,384,029 – 9,384,132 |
| Length | 103 |
| Max. P | 0.954666 |

| Location | 9,384,029 – 9,384,132 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -28.20 |
| Energy contribution | -29.23 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9384029 103 + 20766785 UCCGUUGCAGAUUCAAUUUGCUGAUUGAUUUACCUUGAUGAUGGGACCCUGUCCACUCAUAUAUGGGGGCAAGUUGGAGCUGCCGCCAUUGUGGGC---AUGGGGU ((((((((((..((((((....))))))......))).))))))).((((((((((......((((.((((.((....)))))).)))).))))))---).))).. ( -37.50) >DroPse_CAF1 5749 85 + 1 -------------CAGUU-----UUUGAUUUACCUUGAUGAUGGGACCCUGUCCACUCAUAUAUGGCGGCAAGUUGGAGCUGCCGCCAUUGUGGGC---AUGGGGU -------------.....-----.........(((.......))).((((((((((......(((((((((.((....))))))))))).))))))---).))).. ( -35.20) >DroEre_CAF1 6286 102 + 1 UCUGUUGCAGAUGCUAU-UGCUGAUUGAUUUACCUUGAUGAUGGGACCCUGUCCACUCAUAUAUGGGGGCAAGUUGGAGCUGCCGCCAUUGUGGGC---AUGGGGU ((.(((((((......)-))).))).))....(((.......))).((((((((((......((((.((((.((....)))))).)))).))))))---).))).. ( -33.30) >DroWil_CAF1 8304 92 + 1 UUG------------GUUUUUU--GUGAUUUACCUUUAUGAUGGGACCCUGUCCACUCAUAUAUGGCGGCAAGUUGCCGCUGCCGCCAUUUUGGCCAGAAUUGGGU (((------------(((....--............(((((..((((...))))..))))).(((((((((.((....)))))))))))...))))))........ ( -28.40) >DroAna_CAF1 5853 103 + 1 UUGAUUUCCCUUGAGAUUGGCUGUUUGAUUUACCUUGAUGAUGGGACCCUGUCCACUCAUGUAUGGCGGCAAGUUGGAGCUGCCGCCAUUCUGGGC---AUUGGGU (..(((((....)))))..).............((..(((...((((...)))).((((.(.(((((((((.((....))))))))))).))))))---))..)). ( -33.90) >DroPer_CAF1 7665 85 + 1 -------------CAGUU-----UUUGAUUUACCUUGAUGAUGGGACCCUGUCCACUCAUAUAUGGCGGCAAGUUGGAGCUGCCGCCAUUGUGGGC---AUGGGGU -------------.....-----.........(((.......))).((((((((((......(((((((((.((....))))))))))).))))))---).))).. ( -35.20) >consensus U____________CAAUUUGCU_UUUGAUUUACCUUGAUGAUGGGACCCUGUCCACUCAUAUAUGGCGGCAAGUUGGAGCUGCCGCCAUUGUGGGC___AUGGGGU ................................(((.......)))(((((((...((((((.(((((((((.((....)))))))))))))))))....))))))) (-28.20 = -29.23 + 1.03)

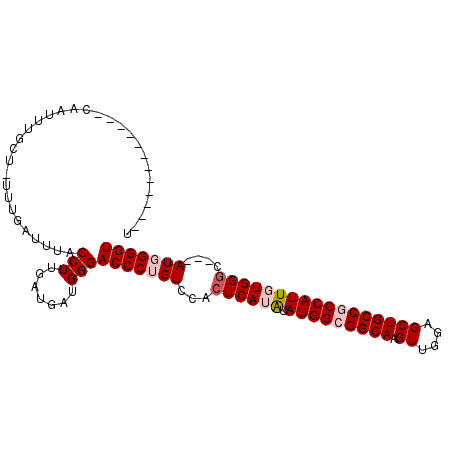
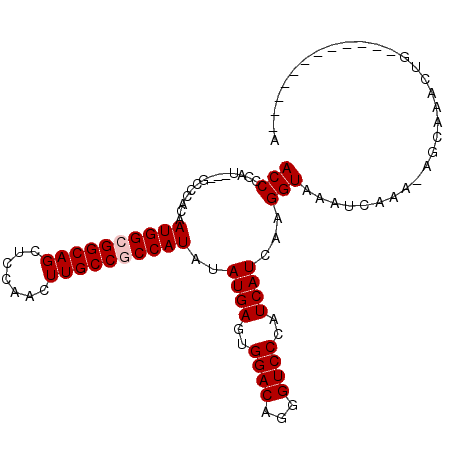
| Location | 9,384,029 – 9,384,132 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9384029 103 - 20766785 ACCCCAU---GCCCACAAUGGCGGCAGCUCCAACUUGCCCCCAUAUAUGAGUGGACAGGGUCCCAUCAUCAAGGUAAAUCAAUCAGCAAAUUGAAUCUGCAACGGA ...((.(---(((....((((.(((((.......))))).))))..((((..((((...))))..))))...))))..(((((......))))).........)). ( -23.80) >DroPse_CAF1 5749 85 - 1 ACCCCAU---GCCCACAAUGGCGGCAGCUCCAACUUGCCGCCAUAUAUGAGUGGACAGGGUCCCAUCAUCAAGGUAAAUCAAA-----AACUG------------- ......(---(((....((((((((((.......))))))))))..((((..((((...))))..))))...)))).......-----.....------------- ( -26.10) >DroEre_CAF1 6286 102 - 1 ACCCCAU---GCCCACAAUGGCGGCAGCUCCAACUUGCCCCCAUAUAUGAGUGGACAGGGUCCCAUCAUCAAGGUAAAUCAAUCAGCA-AUAGCAUCUGCAACAGA ......(---(((....((((.(((((.......))))).))))..((((..((((...))))..))))...)))).........(((-........)))...... ( -22.10) >DroWil_CAF1 8304 92 - 1 ACCCAAUUCUGGCCAAAAUGGCGGCAGCGGCAACUUGCCGCCAUAUAUGAGUGGACAGGGUCCCAUCAUAAAGGUAAAUCAC--AAAAAAC------------CAA ...........(((...((((((((((.(....))))))))))).(((((..((((...))))..)))))..))).......--.......------------... ( -31.40) >DroAna_CAF1 5853 103 - 1 ACCCAAU---GCCCAGAAUGGCGGCAGCUCCAACUUGCCGCCAUACAUGAGUGGACAGGGUCCCAUCAUCAAGGUAAAUCAAACAGCCAAUCUCAAGGGAAAUCAA .(((...---.....(.((((((((((.......)))))))))).).((((.((((...)))).........(((..........)))...)))).)))....... ( -30.30) >DroPer_CAF1 7665 85 - 1 ACCCCAU---GCCCACAAUGGCGGCAGCUCCAACUUGCCGCCAUAUAUGAGUGGACAGGGUCCCAUCAUCAAGGUAAAUCAAA-----AACUG------------- ......(---(((....((((((((((.......))))))))))..((((..((((...))))..))))...)))).......-----.....------------- ( -26.10) >consensus ACCCCAU___GCCCACAAUGGCGGCAGCUCCAACUUGCCGCCAUAUAUGAGUGGACAGGGUCCCAUCAUCAAGGUAAAUCAAA_AGCAAACUG____________A (((..............((((((((((.......))))))))))..((((..((((...))))..))))...)))............................... (-22.25 = -22.58 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:33 2006