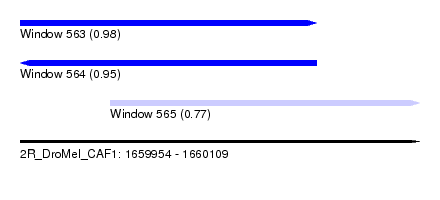
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,659,954 – 1,660,109 |
| Length | 155 |
| Max. P | 0.984128 |

| Location | 1,659,954 – 1,660,069 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.46 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -30.67 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1659954 115 + 20766785 ----AAAAAAUUCAGUAAAAUUGCAUCAAAAUAA-AAAAGUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUUCUA ----..........(((....)))..........-...((..(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))..)). ( -31.60) >DroPse_CAF1 75350 112 + 1 UAACAAGAAGA----AAAAAAUG----UAAAAAAAUUGUGCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCGUUG ...........----........----............((.(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))).)).. ( -31.20) >DroSec_CAF1 45566 108 + 1 ----AAAA--------AAAAUUGCGUCUAAAAAAUCAAAUUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUCGUA ----....--------..........................(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))..... ( -30.80) >DroSim_CAF1 42478 109 + 1 ----AAAAA-------AAAAUUGCAUCUAAAAAAGUAAAUUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUUAUG ----.....-------..........................(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))..... ( -30.80) >DroEre_CAF1 49963 112 + 1 ----AAGUAGU----AAAAAUUGGAUCAAAAAAUUGUUUUUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUGACA ----.......----...................((((....(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))).)))) ( -31.30) >DroPer_CAF1 69326 112 + 1 UAACAAGAAGA----AAAAAAUG----UAAAAAAGUUGUGCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCGUUG ...........----........----............((.(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))).)).. ( -31.20) >consensus ____AAAAAG_____AAAAAUUGCAUCUAAAAAAGUAAAGUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUGAUA ..........................................(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))..... (-30.67 = -30.67 + 0.00)

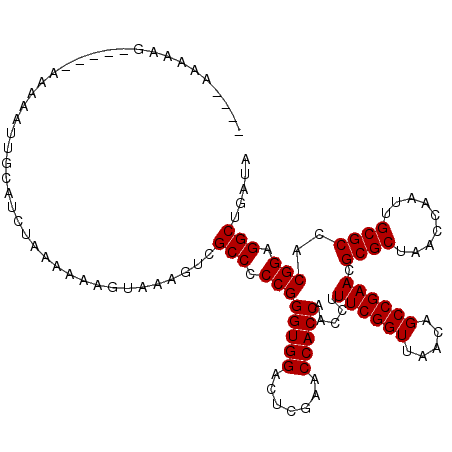
| Location | 1,659,954 – 1,660,069 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.46 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -37.03 |
| Energy contribution | -37.03 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1659954 115 - 20766785 UAGAAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGACUUUU-UUAUUUUGAUGCAAUUUUACUGAAUUUUUU---- .((..(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))..))...-..............................---- ( -37.30) >DroPse_CAF1 75350 112 - 1 CAACGGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGCACAAUUUUUUUA----CAUUUUUU----UCUUCUUGUUA ...(.(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))).).............----........----........... ( -37.50) >DroSec_CAF1 45566 108 - 1 UACGAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGAAUUUGAUUUUUUAGACGCAAUUUU--------UUUU---- .....(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))(..(((((....)))))..)......--------....---- ( -38.30) >DroSim_CAF1 42478 109 - 1 CAUAAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGAAUUUACUUUUUUAGAUGCAAUUUU-------UUUUU---- .....(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))..........................-------.....---- ( -37.10) >DroEre_CAF1 49963 112 - 1 UGUCAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGAAAAACAAUUUUUUGAUCCAAUUUUU----ACUACUU---- .(((((((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))(((((....))))))))).........----.......---- ( -37.30) >DroPer_CAF1 69326 112 - 1 CAACGGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGCACAACUUUUUUA----CAUUUUUU----UCUUCUUGUUA ...(.(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))).).............----........----........... ( -37.50) >consensus CAACAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGAAAUAAAUUUUUUAGAUGCAAUUUUU_____CUUCUU____ .....(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))).......................................... (-37.03 = -37.03 + 0.00)

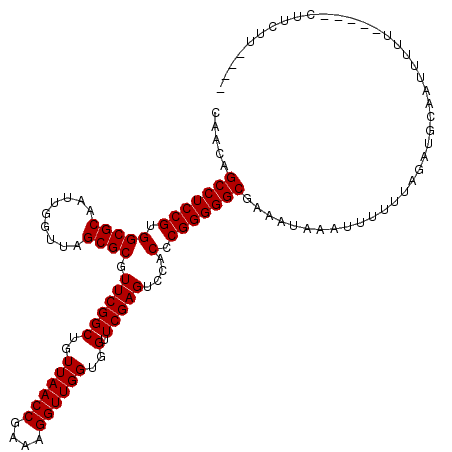
| Location | 1,659,989 – 1,660,109 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.71 |
| Mean single sequence MFE | -38.49 |
| Consensus MFE | -30.68 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1659989 120 + 20766785 UCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUUCUACAUUUUGAUGGACUAAUGUCGAUUGUCAGCCAAGCCAAAA ..(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))...........(((.(((....))).)))................ ( -35.60) >DroPse_CAF1 75382 113 + 1 CCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCGUUGCCGCUUUCACUUCAAAUGGCGAUUAAAAGAGAA------- ..(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))).(((((((.(((......))))))))))..........------- ( -36.60) >DroSec_CAF1 45594 120 + 1 UCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUCGUACAUUCUGUUAGACUAAUGUCGAGUGUAAGCCAAGCCAAAA ..((..((((((((.......))))).....((((((.....)))))).((((.........))))..))).((((..(((((((.....(((....))))))))))))))..))..... ( -39.10) >DroSim_CAF1 42507 120 + 1 UCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUUAUGCAUUUCAUUCGACUAAUGUCGAGUGUAAGCCAAGCCAAAA ......((((((((.......))))).....((((((.....)))))).((((.........))))..))).(((((..((.((.((((((((....)))))))).)))).))))).... ( -44.30) >DroEre_CAF1 49995 120 + 1 UCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUGACAUAAGUUCUUGGUCUAAUGUCAAGGGUAAGCCAAGCCAAAA ..(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))((((((.((........)).))))))..(((.......))).... ( -39.50) >DroPer_CAF1 69358 113 + 1 CCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCGUUGCCGCUUUCACUUCAUAUGGCGAUUAAAAGAGAA------- ..(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))).(((((((.............)))))))..........------- ( -35.82) >consensus UCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUGAUACAUUUUGUUCGACUAAUGUCGAGUGUAAGCCAAGCCAAAA ..(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))...............(((....))).................... (-30.68 = -30.85 + 0.17)

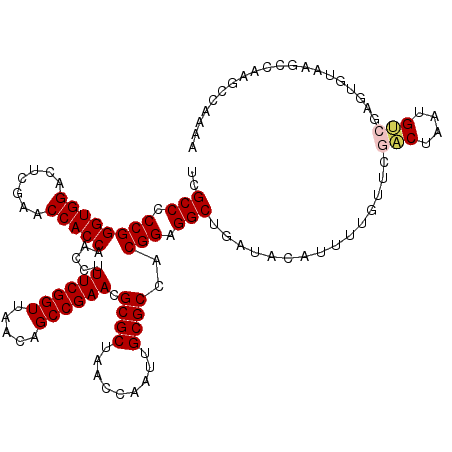
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:02 2006