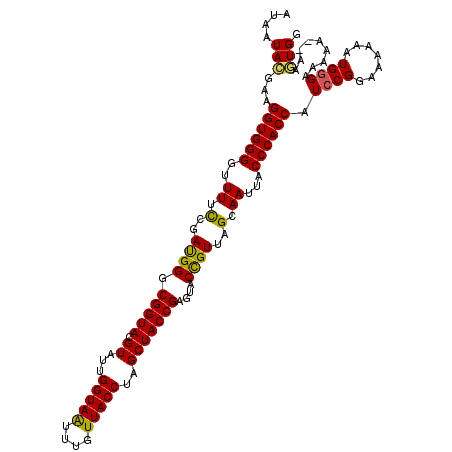
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,344,863 – 9,344,976 |
| Length | 113 |
| Max. P | 0.997521 |

| Location | 9,344,863 – 9,344,976 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -30.66 |
| Energy contribution | -30.02 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9344863 113 + 20766785 AUAAUACGAAGGUGGGGUUUUCCGAUGGGCGGUACGUAUUGGUAAUUUGUUACCUAGCUACCGAGUACCGUUAGCAAUUACCCACCAUCCGGAAAAAAUGGGAAAAA--AAGUGG ....(((...((((((..((.(..((((.(((((.((...(((((....)))))..)))))))....))))..).))...)))))).((((.......)))).....--..))). ( -33.90) >DroSec_CAF1 15052 115 + 1 AUAAUACGAAGGUGGGGUUUUCCCAUGGGCGGUACGUAUUGGUAAUUUGUUACCUAGCUACCGAGUACUGUUAGCAAUUACCCACCAUCCGGAAAAAAUGGGGAAAAAUAAGUGG ....(((....)))...(((((((((((((((((.((...(((((....)))))..))))))).....((....))....))).((....)).....)))))))))......... ( -29.30) >DroSim_CAF1 15630 113 + 1 AUAAUACGAAGGUGGGGUUUUCCGAUGGGCGGUACGUAUUGGUAAUUUGUUACCUAGCUACCGAGUACCGUUAGCAAUUACCCACCAUCCGGAAAAAAUGGGGAAAA--AAGUGG ....(((...((((((..((.(..((((.(((((.((...(((((....)))))..)))))))....))))..).))...)))))).((((.......)))).....--..))). ( -33.90) >DroEre_CAF1 15320 111 + 1 AUUAUAGGAAGGUGGGGUUUUUGGAUGGGCGGUACGUAUUGGUAGUUUGUUACCUAGCUACCGAGUACCGUUAGCAAUUACCCACCAUCCGGAAAAAAUGGGAAA----AACUGC ......(((.((((((....(((....((((((((...(((((((((........)))))))))))))))))..)))...)))))).)))...............----...... ( -40.10) >DroYak_CAF1 18146 111 + 1 AUAAUACGAGGGUGGGGUUUUUGGACGGGCGGUACGUAUUGGUAGUUUGUUACCUAGCUACCGAGUACCGUUAGCAAUUACCCACCAUCCGGAAAAAAUGUGAAA----AACUGG ......((..((((((....(((....((((((((...(((((((((........)))))))))))))))))..)))...))))))...))..............----...... ( -36.60) >consensus AUAAUACGAAGGUGGGGUUUUCCGAUGGGCGGUACGUAUUGGUAAUUUGUUACCUAGCUACCGAGUACCGUUAGCAAUUACCCACCAUCCGGAAAAAAUGGGAAAAA__AAGUGG ....(((...((((((..((.(..((((.(((((.((...(((((....)))))..)))))))....))))..).))...)))))).((((.......)))).........))). (-30.66 = -30.02 + -0.64)


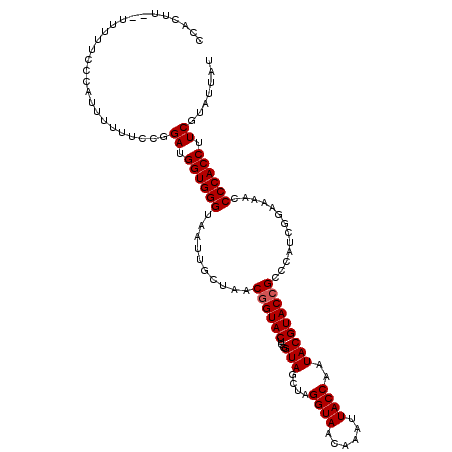
| Location | 9,344,863 – 9,344,976 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9344863 113 - 20766785 CCACUU--UUUUUCCCAUUUUUUCCGGAUGGUGGGUAAUUGCUAACGGUACUCGGUAGCUAGGUAACAAAUUACCAAUACGUACCGCCCAUCGGAAAACCCCACCUUCGUAUUAU ......--...........(((((((.((((..((((...((((.((.....)).))))..(((((....)))))......))))..)))))))))))................. ( -27.30) >DroSec_CAF1 15052 115 - 1 CCACUUAUUUUUCCCCAUUUUUUCCGGAUGGUGGGUAAUUGCUAACAGUACUCGGUAGCUAGGUAACAAAUUACCAAUACGUACCGCCCAUGGGAAAACCCCACCUUCGUAUUAU ........((((((((((((.....)))))((((((....((((...(....)..))))..(((((....)))))..........)))))))))))))................. ( -25.50) >DroSim_CAF1 15630 113 - 1 CCACUU--UUUUCCCCAUUUUUUCCGGAUGGUGGGUAAUUGCUAACGGUACUCGGUAGCUAGGUAACAAAUUACCAAUACGUACCGCCCAUCGGAAAACCCCACCUUCGUAUUAU ......--...........(((((((.((((..((((...((((.((.....)).))))..(((((....)))))......))))..)))))))))))................. ( -27.30) >DroEre_CAF1 15320 111 - 1 GCAGUU----UUUCCCAUUUUUUCCGGAUGGUGGGUAAUUGCUAACGGUACUCGGUAGCUAGGUAACAAACUACCAAUACGUACCGCCCAUCCAAAAACCCCACCUUCCUAUAAU ......----...............(((.((((((...(((....((((((..(((((...(....)...))))).....))))))......)))....)))))).)))...... ( -27.80) >DroYak_CAF1 18146 111 - 1 CCAGUU----UUUCACAUUUUUUCCGGAUGGUGGGUAAUUGCUAACGGUACUCGGUAGCUAGGUAACAAACUACCAAUACGUACCGCCCGUCCAAAAACCCCACCCUCGUAUUAU ......----................((.((((((...(((...((((....(((((....((((......))))......))))).)))).)))....)))))).))....... ( -24.90) >consensus CCACUU__UUUUUCCCAUUUUUUCCGGAUGGUGGGUAAUUGCUAACGGUACUCGGUAGCUAGGUAACAAAUUACCAAUACGUACCGCCCAUCGGAAAACCCCACCUUCGUAUUAU ..........................((.((((((..........((((((...(((....((((......))))..))))))))).............)))))).))....... (-21.66 = -21.86 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:21 2006