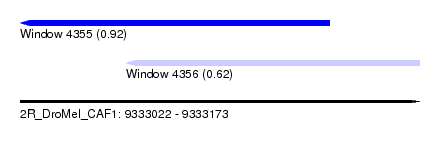
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,333,022 – 9,333,173 |
| Length | 151 |
| Max. P | 0.919625 |

| Location | 9,333,022 – 9,333,139 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.37 |
| Mean single sequence MFE | -39.53 |
| Consensus MFE | -15.40 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
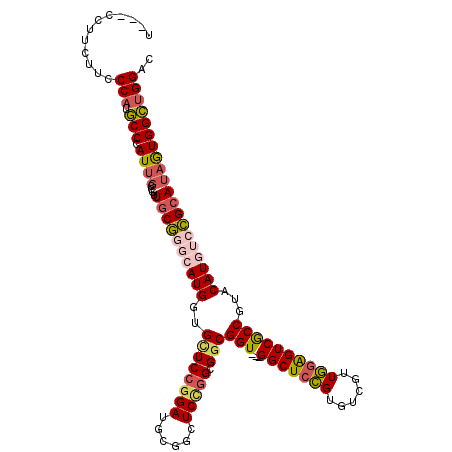
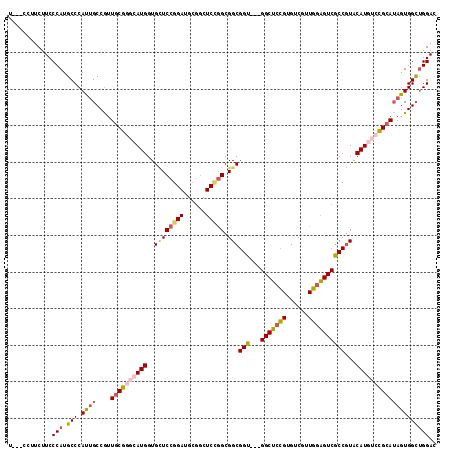
>2R_DroMel_CAF1 9333022 117 - 20766785 GGUGAUCCGGAUGCGGCUCCGGCGGCGGU---GGCUCCGUGUCGUUGGAGUCACCAUACAUAUCCGCAUAGUGGCUGGACACACCGGGCGCCCACUGCAACAGUUGUACAAUAUACCACA ((((....((((((.((....)).))(((---(((((((......))))))))))......))))(((..(((.((((.....)))).)))....)))...............))))... ( -45.50) >DroVir_CAF1 4231 102 - 1 CUGGCUCCGG------CUCC------------GGUUCUGUGCCAUUCACAUCCAUAUACACAUCAUUGUAAUGGGCCGUUACGCCGGGCGCCAGUCGCAUCAGUUGUACAAUAUACCACA ((((((((((------(..(------------(((..((((.....)))).((((.((((......))))))))))))....)))))).)))))........((.(((.....))).)). ( -32.20) >DroPse_CAF1 3707 102 - 1 GGUGCUCUGG------CUCC------------GGCUCAGUGCCGUUCAUGUCGCUGUACAUUUCAGCGUAGUGGGCGGACACCCCGGGCGCCAGUUGCAUCAGUUGCACAAUAUACCACA (((((.((((------((((------------((....(((((((((((..(((((.......)))))..)))))))).))).))))).)))))..)))))................... ( -46.60) >DroWil_CAF1 4478 99 - 1 CAUGCUCUGG------CUCC------------GGUUCGGUGCCAUUCAUAUCAAGAUACAUUUCACUAUAG---GCCGACACUCCGGGUGUCCAAUGCACUAGUUGCACAAUAUACCACA ..(((..(((------(.((------------(...))).))))....................((((...---((.(((((.....)))))....))..)))).)))............ ( -20.50) >DroAna_CAF1 3336 108 - 1 GGUGUUCUG------------GCGGCGGUCUCGGCUCGGUGCCGUUCAGGUCGCCGUGCAUAUCGGCAAAAUGGGCGGACACUCCGGGCGCCCAUUGCAUCAGUUGCACAAUAUACCACA (((((..((------------((((((..((((((.....))))...))..))))))(((.....(((..(((((((..(.....)..))))))))))......))).))..)))))... ( -49.00) >DroPer_CAF1 3682 102 - 1 GGUGCUCUGG------CUCC------------GGCUCAAUGCCGUUCAUGUCGCUGUACAUUUCAGCGUAGUGGGCGGACACCCCGGGCGCCAGUUGCAUCAGUUGCACAAUAUACCACA (((((.((((------((((------------((.......((((((((..(((((.......)))))..)))))))).....))))).)))))..)))))................... ( -43.40) >consensus GGUGCUCUGG______CUCC____________GGCUCAGUGCCGUUCAUGUCGCUAUACAUAUCAGCAUAGUGGGCGGACACCCCGGGCGCCAAUUGCAUCAGUUGCACAAUAUACCACA (((((.((((......................(((.....)))..........((((.(((.........))).)))).....)))))))))...((((.....))))............ (-15.40 = -16.18 + 0.78)

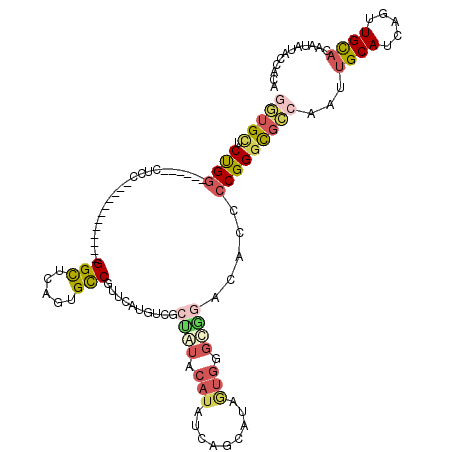
| Location | 9,333,062 – 9,333,173 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -50.28 |
| Consensus MFE | -38.67 |
| Energy contribution | -39.62 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
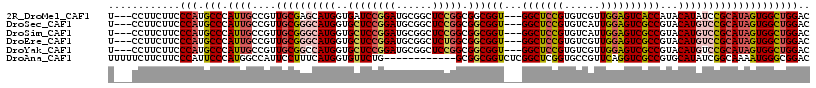
>2R_DroMel_CAF1 9333062 111 - 20766785 U---CCUUCUUCCCAUGCCCAUUGCCGUUGCGAGCAUGGUGAUCCGGAUGCGGCUCCGGCGGCGGU---GGCUCCGUGUCGUUGGAGUCACCAUACAUAUCCGCAUAGUGGCUGGAC .---........(((.((((...(((((((.((((.(((....)))......)))))))))))(((---(((((((......))))))))))...............).)))))).. ( -47.10) >DroSec_CAF1 3353 111 - 1 U---CCUUCUUCCCAUGCCCAUUGCCGUUGCGGGCAUGGUGCUCCGGAUGCGGCUCCGGCGGCGGU---GGCUCCGUGUCAUUGGAGUCGCCGUACAUGUCCGCAUAGUGGCUGGAC .---........(((.(((.((((....((((((((((...(.(((((......))))).)(((((---(((((((......)))))))))))).)))))))))))))))))))).. ( -55.60) >DroSim_CAF1 3377 111 - 1 U---CCUUCUUCCCAUGCCCAUUGCCGUUGCGGGCAUGGUGCUCCGGAUGCGGCUCCGGCGGCGGU---GGCUCCGUGUCAUUGGAGUCGCCGUACAUGUCCGCAUAGUGGCUGGAC .---........(((.(((.((((....((((((((((...(.(((((......))))).)(((((---(((((((......)))))))))))).)))))))))))))))))))).. ( -55.60) >DroEre_CAF1 3459 111 - 1 U---CCUUCUUCCCAUGCCCAUUGCCGUUGCGGGCAUGGUGCUCCGGAUGCGGCUCUGGCGGCGGU---GGCUCCGUGUCGUUGGAGUCGCCGUACAUGUCCGCAUAGUGGCUGGAC (---((......((((((((...((....)))))))))).((..(..((((((...((...(((((---(((((((......)))))))))))).))...)))))).)..)).))). ( -53.70) >DroYak_CAF1 6123 111 - 1 U---CCUUCUUCCCAUGCCCAUUGCCGUUGCGGCCAUGGUGCUCCGGAUGCGGCUCCGGCGGCGGU---GGCUCCGUGUCGUUGGAGUCGCCGUACAUGUCCGCAUAGUGGCUGGAC .---........(((.(((.((((....(((((.((((...(.(((((......))))).)(((((---(((((((......)))))))))))).)))).))))))))))))))).. ( -51.20) >DroAna_CAF1 3376 105 - 1 UUUUUCUUCUUCCCAUUCCCAUGGCCAUUCCUUUCAUGGUGUUCUG------------GCGGCGGUCUCGGCUCGGUGCCGUUCAGGUCGCCGUGCAUAUCGGCAAAAUGGGCGGAC ..........(((....(((((.(((...((......)).......------------((((((..((((((.....))))...))..)))))).......)))...))))).))). ( -38.50) >consensus U___CCUUCUUCCCAUGCCCAUUGCCGUUGCGGGCAUGGUGCUCCGGAUGCGGCUCCGGCGGCGGU___GGCUCCGUGUCGUUGGAGUCGCCGUACAUGUCCGCAUAGUGGCUGGAC ............(((.(((.((((....((((((((((..((((((((......))))).)))(((...(((((((......))))))))))...)))))))))))))))))))).. (-38.67 = -39.62 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:17 2006