| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,332,087 – 9,332,200 |
| Length | 113 |
| Max. P | 0.939871 |

| Location | 9,332,087 – 9,332,200 |
|---|---|
| Length | 113 |
| Sequences | 6 |
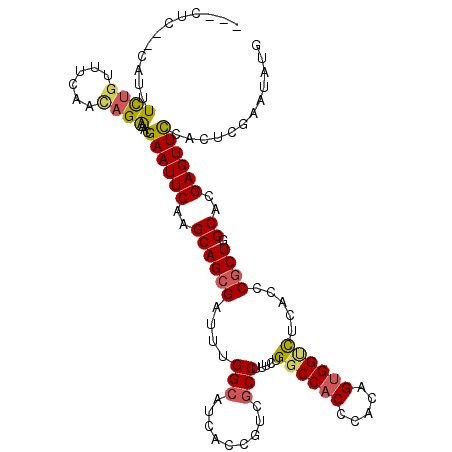
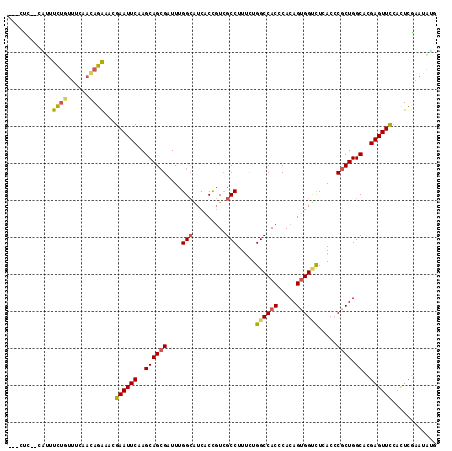
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.41 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.75 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9332087 113 + 20766785 ---CUCAAUGCAUCUAUUUGCACAGAAACGAAUUCAAGCAGCGUUUUGGCGUCACCGUCGCCUUUUUGGCCACCCACAGUGGUCUCACCCGCUGGCAUGAGUUCCACUCGAAUGCU ---.((..((((......))))..))...(((((((.((((((....((((.......)))).....((((((.....)))))).....)))).)).)))))))............ ( -34.10) >DroVir_CAF1 2771 114 + 1 UAUAUC--GAUUUCUGUUCCAACAGACACGAAUUCAAGCAGCGAUUUGGCAUCACCGUCGCCUUUCUGGCCACGCACAGUGGGCUAACACGCUGGCAUGAGUUUCAUUCGAAUAUG ....((--((..(((((....)))))...(((((((.((((((..(((((.(((((((.(((.....))).)))....)))))))))..)))).)).)))))))...))))..... ( -36.40) >DroPse_CAF1 2782 106 + 1 ---CCCU-UUUAUCUGUUGGCUUAGAAACGAAUUCAAGCAGCGUUUUGGCGUCACCGUCGCCUUCCUGGCCACCCACAGCGGCCUCACACGCUGGCACGAGUUCAACUUG------ ---....-....((((......))))...((((((..(((((((...((((.......)))).....((((.(.....).))))....))))).))..))))))......------ ( -29.90) >DroGri_CAF1 2798 114 + 1 UAUCCU--CGUUUCUGUUUUAACAGACACGAAUUCAAGCAGAGAUUUGGGAUCACCGUCGCCUUUUUGGCCACGCACAGUGGUUUGACCCGCUGGCACGAGUUCCACUCGAAUAUG .....(--(((.(((((....))))).)))).........(((...(((((((...((.(((.....))).))((.((((((......))))))))..)).))))))))....... ( -35.30) >DroMoj_CAF1 2507 114 + 1 UAUCUC--GAUUUCUGUUCCAACAGACACGAAUUCAAGCAGCGCUUUGGCAUCACCGUCGCCUUUCUGGCCACGCACAGUGGUCUGACCCGCUGGCACGAGUUCCACUCGAACAUG ....((--((..(((((....)))))...((((((..((((((..(..(.((((((((.(((.....))).)))....))))))..)..)))).))..))))))...))))..... ( -34.40) >DroAna_CAF1 2436 113 + 1 ---CGAUUCCCUUUCGUUUAAACAGGAACGAAUUCAAGCAGCGCUUCGGCGUCACUUUGGCCUUUCUGGCCACCCACAGUGGCCUCACCCGCUGGCACGAGUUCCAUUCCAACGGA ---.........((((((......((((.(((((((((..((((....))))..)))(((((.....)))))....((((((......))))))....))))))..)))))))))) ( -37.60) >consensus ___CUC__CAUUUCUGUUUCAACAGAAACGAAUUCAAGCAGCGAUUUGGCAUCACCGUCGCCUUUCUGGCCACCCACAGUGGUCUCACCCGCUGGCACGAGUUCCACUCGAAUAUG ............((((......))))...((((((..((((((....(((.........))).....((((((.....)))))).....)))).))..))))))............ (-23.58 = -23.75 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:14 2006