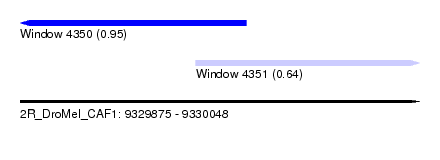
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,329,875 – 9,330,048 |
| Length | 173 |
| Max. P | 0.948841 |

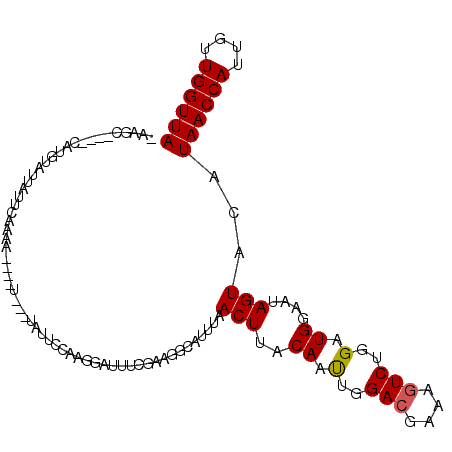
| Location | 9,329,875 – 9,329,973 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.99 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -14.99 |
| Energy contribution | -15.02 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9329875 98 - 20766785 -AAGC----CAUGUAUGACACAAAA---------GAUCCACAGAUUUUGAAGGCAUUUAACUUACAAUUGGACGAAAGUCUGGAUGGAAUAGUACAUAACCAUUGUUGGUUA -..((----(.(((.....))).((---------((((....))))))...))).....(((..((.(..(((....)))..).))....)))...((((((....)))))) ( -23.80) >DroVir_CAF1 172 110 - 1 AAAGGUAUACAUAUAGUGUUGAAAA-GUGUAAAUAUUUCUGGUAUUUUGUUG-UCUAGUACUUACAAUUGGACGAAAAUCUGGAUGGAAUAGUACAUAACCAUUGUUGGUUA ......((((.....))))......-(((((......((..(.((((..(((-((((((.......))))))))))))))..))........)))))(((((....))))). ( -20.04) >DroSec_CAF1 174 99 - 1 -AAGA----CAAGGAUUAUUUAAAA----U---AAUUCCUACUAUUUCGAUGGCAUUC-ACUUACAAUUGGACGAAAGUCUGGAUGGAAUAGUACAUAACCAUUGUUGGUUA -....----...(((((((.....)----)---))))).(((((((((((......))-........(..(((....)))..)..)))))))))..((((((....)))))) ( -25.30) >DroSim_CAF1 196 98 - 1 -AAGC----CAUGUAUUACCCAAAA---------GUUCCAAAGAUUUUGAAGGCAUUUAACUUACAAUUGGACGAAAGUCUGGAUGGAAUAGUACAUAACCAUUGUUGGUUA -....----.(((((((........---------((((((......((((((........))).)))(..(((....)))..).)))))))))))))(((((....))))). ( -24.00) >DroEre_CAF1 178 105 - 1 AAAGC----CAAGAAUUAUUCCAAGCCUUU---UAUGCCAAGCAUUUCGAAGGGAUGUCACUUACAAUUGGACGAAAGUCUGGAUGGAAUAGUACAUAACCAUUGUUGGUUA .....----...(.(((((((((..(((((---.((((...))))...)))))..............(..(((....)))..).))))))))).).((((((....)))))) ( -29.90) >DroYak_CAF1 2883 104 - 1 AAAGA----AAUGGAUUAUUGAAAACGUUU---UAUGCCUAGGAUUUCGA-GACAUUUCACUUACAACUGGACGAAAGUCUGGAUGGAAUAGUACAUAACCAUUGUUGGUUA ...((----((((.....(((((...((((---((....)))))))))))-..))))))(((..((.(..(((....)))..).))....)))...((((((....)))))) ( -26.10) >consensus _AAGC____CAUGUAUUAUUCAAAA____U___UAUUCCAAGGAUUUCGAAGGCAUUUAACUUACAAUUGGACGAAAGUCUGGAUGGAAUAGUACAUAACCAUUGUUGGUUA ...........................................................(((..((.(..(((....)))..).))....)))...((((((....)))))) (-14.99 = -15.02 + 0.03)


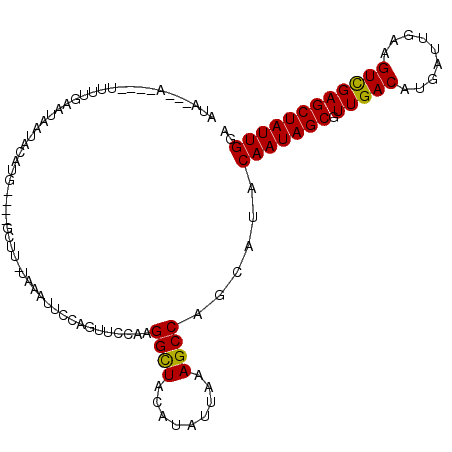
| Location | 9,329,951 – 9,330,048 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.33 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9329951 97 + 20766785 UC---------UUUUGUGUCAUACAUG----GCUU-UAAAUUCCAGUUCCAAGGCUACAUAUUAAAGCCAGCAUACAAUAGCGUUGACAUGAUUGAAGUCGAGCUAUUGGA ..---------...(((((......((----((((-(((.....(((......))).....))))))))))))))(((((((.(((((.........)))))))))))).. ( -26.00) >DroVir_CAF1 247 110 + 1 AUAUUUACAC-UUUUCAACACUAUAUGUAUACCUUUUGCUGAGCAGUUCCAAGGUUAUAUACUAAAGCCAGCUUACAAUAGCGUUGACAUGAUUGAAGUCGAGCUAUUGGA ..........-............((((((.(((((..(((....)))...)))))))))))...(((....))).(((((((.(((((.........)))))))))))).. ( -22.90) >DroSec_CAF1 249 99 + 1 AUU---A----UUUUAAAUAAUCCUUG----UCUU-UGAAUUUCAGUUCCAAGGCUACAUAUUGAAGCCAGCAUACAAUAGCGUUGACAUGAUUGAAGUCGAGCUAUUGGA ...---.----...............(----((((-.((((....)))).)))))....................(((((((.(((((.........)))))))))))).. ( -20.20) >DroSim_CAF1 272 97 + 1 AC---------UUUUGGGUAAUACAUG----GCUU-UAAAUUCCAGUUCCAAGGCUACAUAUUAAAGCCAGCAUACAAUAGCGUUGACAUGAUUGAAGUUGAGCUAUUGGA ..---------..............((----((((-(((.....(((......))).....))))))))).....(((((((.(..((.........))..)))))))).. ( -22.60) >DroEre_CAF1 254 104 + 1 AUA---AAAGGCUUGGAAUAAUUCUUG----GCUUUUCAAUUCCAGUUCCAAGGCUACAUAUUAAAGCCAGCAUACAAUAGCGUUGACAUGAUUGAAGUCGAGCUAUUGGA ...---....(((((((((.....(((----(....)))).....)))))))((((.........)))).))...(((((((.(((((.........)))))))))))).. ( -26.70) >DroYak_CAF1 2958 104 + 1 AUA---AAACGUUUUCAAUAAUCCAUU----UCUUUUCGAGUACAGUUCCAAGGCUACAUAUUAAAGCCAGCAUACAAUAGCGUUGACAUGAUUGAAGUCGAGCUAUUGGA ...---.......(((((((.((.(((----((...(((.((.(((......((((.........)))).((........)).))))).)))..))))).))..))))))) ( -18.70) >consensus AUA___A____UUUUGAAUAAUACAUG____GCUU_UAAAUUCCAGUUCCAAGGCUACAUAUUAAAGCCAGCAUACAAUAGCGUUGACAUGAUUGAAGUCGAGCUAUUGGA ....................................................((((.........))))......(((((((.(((((.........)))))))))))).. (-17.61 = -17.33 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:12 2006