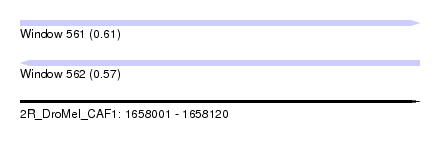
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,658,001 – 1,658,120 |
| Length | 119 |
| Max. P | 0.610160 |

| Location | 1,658,001 – 1,658,120 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -16.83 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.36 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
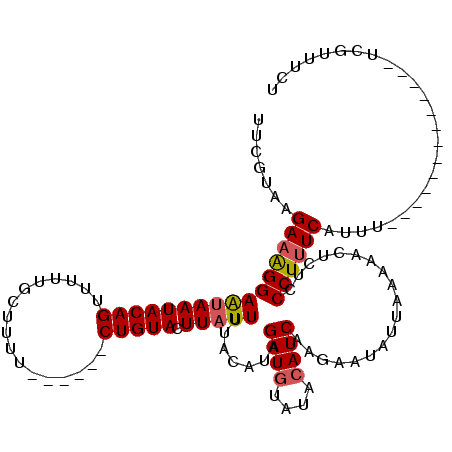
>2R_DroMel_CAF1 1658001 119 + 20766785 UUCGUAAGA-GGGAGAAAUACAGUUUUUGCUUUUUUUGCUCUGUACUUAUUUUACAUAGAUUUAUACAUCAAGAAUAUUAAAUACCCUCCCUUUCAUUUCACAAUUAAUAAUUAAAUUCU ......(((-(((((...(((((.....((.......)).))))).............(((......)))................)))))))).......................... ( -16.80) >DroSec_CAF1 43852 101 + 1 UUCGUAAGAAAGGAAUAAUACAGUUUUUGCUUUU------CUGUACUUAUUUUACAUAGAUGUAUACAUCAAGAAUAUUAAAAACUCUCCCUUUCAUUU-------------UCGUUUCU .......((((((........((((((((...((------(((((.......)))...((((....)))).))))...))))))))...))))))....-------------........ ( -16.50) >DroSim_CAF1 40733 101 + 1 UUCGUAAGAAAGGAAUAAUACAGUUUUUGCUUUU------CUGUACUUAUUUUACUUAGAUGUAUACAUCAAGAAUAUUAAAAACUCUCCCUUUCAUUU-------------UCGUUUCU .......((((((((((((((((...........------))))).)))))...(((.((((....)))))))................))))))....-------------........ ( -17.20) >consensus UUCGUAAGAAAGGAAUAAUACAGUUUUUGCUUUU______CUGUACUUAUUUUACAUAGAUGUAUACAUCAAGAAUAUUAAAAACUCUCCCUUUCAUUU_____________UCGUUUCU .......((((((((((((((((.................))))).))))).......((((....))))...................))))))......................... (-11.80 = -12.36 + 0.56)

| Location | 1,658,001 – 1,658,120 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -13.05 |
| Energy contribution | -13.83 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
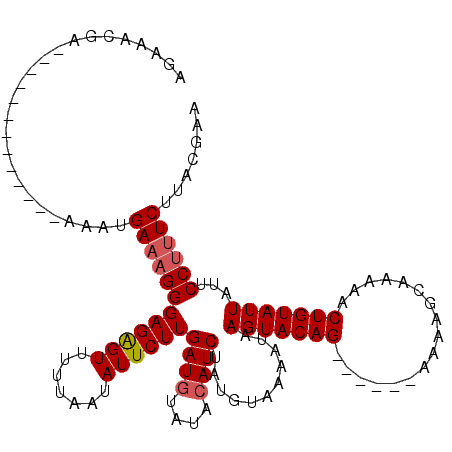
>2R_DroMel_CAF1 1658001 119 - 20766785 AGAAUUUAAUUAUUAAUUGUGAAAUGAAAGGGAGGGUAUUUAAUAUUCUUGAUGUAUAAAUCUAUGUAAAAUAAGUACAGAGCAAAAAAAGCAAAAACUGUAUUUCUCCC-UCUUACGAA ................((((((......(((((((.(((((.((((....(((......))).)))).)))))(((((((.((.......)).....)))))))))))))-).)))))). ( -20.30) >DroSec_CAF1 43852 101 - 1 AGAAACGA-------------AAAUGAAAGGGAGAGUUUUUAAUAUUCUUGAUGUAUACAUCUAUGUAAAAUAAGUACAG------AAAAGCAAAAACUGUAUUAUUCCUUUCUUACGAA .....((.-------------....((((((((((((.......))))))((((....))))...........(((((((------...........)))))))...))))))...)).. ( -18.50) >DroSim_CAF1 40733 101 - 1 AGAAACGA-------------AAAUGAAAGGGAGAGUUUUUAAUAUUCUUGAUGUAUACAUCUAAGUAAAAUAAGUACAG------AAAAGCAAAAACUGUAUUAUUCCUUUCUUACGAA .....((.-------------....((((((((((((.......))))))((((....))))...........(((((((------...........)))))))...))))))...)).. ( -18.50) >consensus AGAAACGA_____________AAAUGAAAGGGAGAGUUUUUAAUAUUCUUGAUGUAUACAUCUAUGUAAAAUAAGUACAG______AAAAGCAAAAACUGUAUUAUUCCUUUCUUACGAA .........................((((((((((((.......))))))((((....))))...........(((((((.................)))))))...))))))....... (-13.05 = -13.83 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:59 2006