
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,327,410 – 9,327,514 |
| Length | 104 |
| Max. P | 0.734694 |

| Location | 9,327,410 – 9,327,514 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -15.33 |
| Consensus MFE | -10.31 |
| Energy contribution | -10.08 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9327410 104 + 20766785 AUCCAUAUUUAACCGAUAUA-----UACUAUAACUAACUAUACUAU--GUAUAACUCGUAUAUAAA--UCUAGCUA-AUAUGGUCUAGGACCAUG-GCAUUCU-UUAGCAACCU--UU ....((((.......(((((-----((.((((......)))).)))--)))).......))))...--....((((-((((((((...)))))))-.......-))))).....--.. ( -16.25) >DroVir_CAF1 1448 101 + 1 UUCCAUAUUUAACCGAUAUA----CUUAUAUAAAUAACUAUACC--------UACUCGUAUAUAAC--UCUAGCUA-ACAUGGUCUAGGACCAUG-AUGUUUUAUUAGCAAACUU-UU .....((((((....(((..----..))).))))))..(((((.--------.....)))))....--....((((-((((((((...)))))))-........)))))......-.. ( -13.90) >DroGri_CAF1 1630 103 + 1 UUCCAUAUUUAACCGAUAUA---CUUAAUAUAACUAAUUAUACC--------UACUCGUAUAUAAC--ACUAGCUA-ACAUGGUCUAGGACCAUG-CUAUUUUAUUAGCAAAUUUUUU ...............(((((---(....(((((....)))))..--------.....))))))...--....((((-((((((((...)))))))-........)))))......... ( -13.80) >DroEre_CAF1 1562 100 + 1 AUCCAUAUUUAACCGAUAUA-----UAAUAUAACUAACUAUACUA------UAACUCGUAUAUAAA--UCUAGCUA-AUAUGGUCUAGGACCAUG-GCAUUCU-UUAGCAACCU--UU .............(((..((-----((.((((......)))).))------))..)))........--....((((-((((((((...)))))))-.......-))))).....--.. ( -13.81) >DroAna_CAF1 1583 106 + 1 AUCCAUAUUUAACCGAUAUAUCGUAUACUCUAACUAACUAUACUA------UAACUCGUAUAUAAA--UCUAGCUAAACAUGGUCUAGGACCAUG-GCAUUCU-UUAGCAACCU--UU .............(((..(((.(((((...........))))).)------))..)))........--....(((((((((((((...)))))))-......)-))))).....--.. ( -16.50) >DroPer_CAF1 1575 112 + 1 AUCCAUAUUUAACCAAAAUA---UAUCCAAUAACUAACUAUUAUAGUACUAUAACUAGUAUAUAAAACUCUAGCUAAACAUGGUCUAGGACCAUGGGCAUUCU-UUAGCAACAU--UU ....((((((.....)))))---)................(((((.(((((....)))))))))).......(((((((((((((...))))))).......)-))))).....--.. ( -17.71) >consensus AUCCAUAUUUAACCGAUAUA_____UAAUAUAACUAACUAUACUA______UAACUCGUAUAUAAA__UCUAGCUA_ACAUGGUCUAGGACCAUG_GCAUUCU_UUAGCAACCU__UU ........................................................................((((..(((((((...)))))))..........))))......... (-10.31 = -10.08 + -0.22)

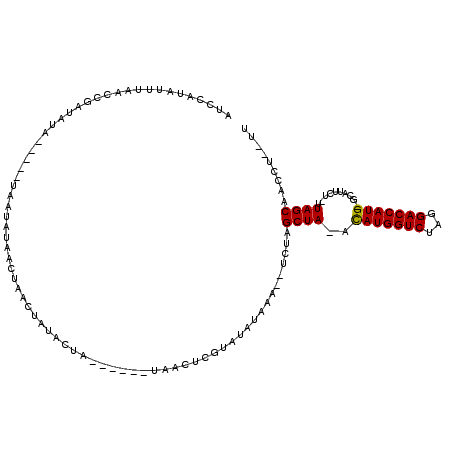
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:10 2006