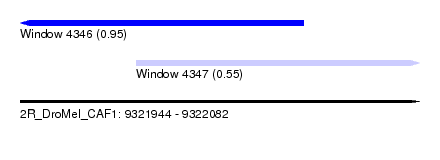
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,321,944 – 9,322,082 |
| Length | 138 |
| Max. P | 0.953436 |

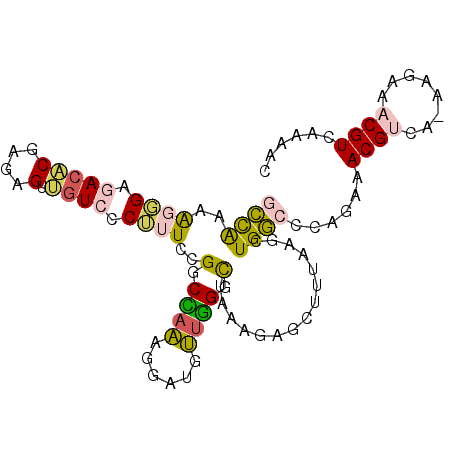
| Location | 9,321,944 – 9,322,042 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.91 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -19.38 |
| Energy contribution | -19.85 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9321944 98 - 20766785 CUCUUUUGCCCAACAUA--------CCUUAGCCGAAAAGGGACAACCCUUGUCUCUCGCUUUUGGCCAUUUGCCGCACUCUUCUUUCCACCCAGUGCGCUUGCCCU .......((........--------.....((((((((((((((.....)))))))...))))))).....((.(((((.............)))))))..))... ( -21.82) >DroSec_CAF1 206621 98 - 1 CUCUUUAACCCAACAUC--------CUUUGGCCGGAAAGGGACAACUCUCGUGUCUCCCUUUUGGCCAUUUGCCGCACUCUUCUGGCCGCUCAGUGCGCGUGCCCU .................--------...((((((.(((((((..((....))...)))))))))))))...(((((.(....((((....)))).).))).))... ( -27.90) >DroSim_CAF1 193922 98 - 1 CUCUUUAACCCAACAUC--------CUUUGGCCGGAAAGGGACAACUAUCGUGUCUCCCUUUUGGCCAUUUGCCGCACUCUUCUGGCCGCCCAGUGCGCGUGCCCU .................--------...((((((.(((((((..((....))...)))))))))))))...(((((.(....((((....)))).).))).))... ( -30.20) >DroEre_CAF1 191904 106 - 1 CUCUGUCGCCCAACAUCCCAAUCUCCCUUGGCCGGAAAGGGAUAACCCUCGAGUCCUCCUUUCGGCCAUUCGCCGCACUCUUCUGGGCGUCCAGUGCGCCUGCCCU ..(((.((((((............(...(((((((((.(((((.........)))))..)))))))))...)...........))))))..)))............ ( -29.40) >DroYak_CAF1 191208 98 - 1 CUCUGUAGCCCCACAUC--------CCUUGGCCGAAAAGGGACAACCCUCGAGUCUCCCUUUCGGCCAUUCGCCGCACUCUUCUGGGCGCCAAGUGCGCCUGUCCU ...((..((........--------...(((((((((.((((..((......)).)))))))))))))...))..)).......((((((.....))))))..... ( -31.94) >DroPer_CAF1 169312 97 - 1 UUCAU-CGCCUAGUUGC--------CACUAGCCAGAGCGAGACCACAGCCGGGUUCCAGCAUUAAGCUCUAGCUGCACUGCUCUGGGCUACCACUGCGCCUGCCCU .((.(-(((((.((((.--------...)))).)).))))))........((((....(((...(((((.(((......)))..))))).....)))....)))). ( -27.60) >consensus CUCUUUAGCCCAACAUC________CCUUGGCCGGAAAGGGACAACCCUCGUGUCUCCCUUUUGGCCAUUUGCCGCACUCUUCUGGCCGCCCAGUGCGCCUGCCCU ............................(((((((((.((((((.......))))))..)))))))))...((.(((((.............)))))))....... (-19.38 = -19.85 + 0.48)

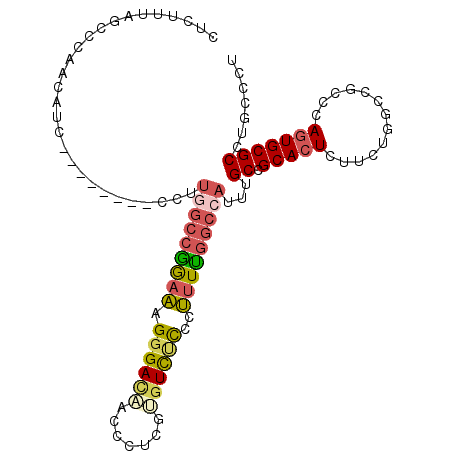
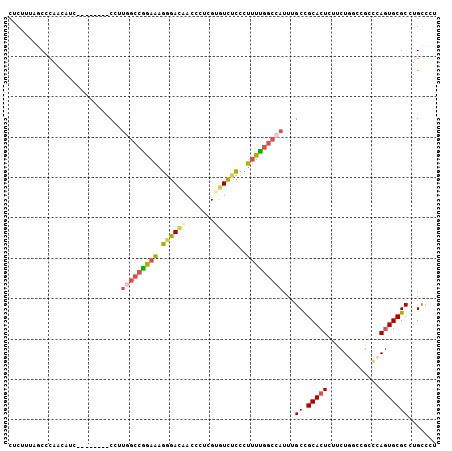
| Location | 9,321,984 – 9,322,082 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 73.19 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -8.86 |
| Energy contribution | -8.03 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9321984 98 + 20766785 GCCAAAAGCGAGAGACAAGGGUUGUCCCUUUUCGGCUAAGGUAUGUUGGGCAAAAGAGCUUUAAGCUGGCCCAGAAACGUUAAAAGAAACGUCAAAAC (((...(((..((((..((((....)))))))).)))..)))...((((((.....(((.....))).))))))..(((((......)))))...... ( -26.70) >DroPse_CAF1 168918 96 + 1 CUUAAUGCUGGAACCCGGCUGUGGUCUCGCUCUGGCUAGUGGCAACUAGGCG-AUGAACUCAUCGCUGACACAGAAACGCCA-CAGAAACGUCAGAAC .......((((....((.(((((((.....((((.(((((....)))))(((-(((....)))))).....))))...))))-)))...))))))... ( -32.30) >DroSec_CAF1 206661 98 + 1 GCCAAAAGGGAGACACGAGAGUUGUCCCUUUCCGGCCAAAGGAUGUUGGGUUAAAGAGCUUUAAGCUGGCCCAGAAACGUCAAAAGAAACGUCAAAAC (((.(((((((.(.((....))).)))))))..))).....(((((((((((....(((.....)))))))))...)))))................. ( -29.80) >DroSim_CAF1 193962 98 + 1 GCCAAAAGGGAGACACGAUAGUUGUCCCUUUCCGGCCAAAGGAUGUUGGGUUAAAGAGCUUUAAGCUGGCCCAGAAACGUCAAAAGAAACGUCAAAAC (((.((((((((((......))).)))))))..))).....(((((((((((....(((.....)))))))))...)))))................. ( -27.90) >DroYak_CAF1 191248 97 + 1 GCCGAAAGGGAGACUCGAGGGUUGUCCCUUUUCGGCCAAGGGAUGUGGGGCUACAGAGCUUUAAGCUGGCCCAGAAACGUCA-AAGAAACGUCAAAAC (((((((((((((((....)))).)))).))))))).....(((((.(((((....(((.....))))))))....))))).-............... ( -36.30) >DroPer_CAF1 169352 96 + 1 CUUAAUGCUGGAACCCGGCUGUGGUCUCGCUCUGGCUAGUGGCAACUAGGCG-AUGAACUCAUCGCUGACACAGAAACGCCA-CAGAAACGUCAGAAC .......((((....((.(((((((.....((((.(((((....)))))(((-(((....)))))).....))))...))))-)))...))))))... ( -32.30) >consensus GCCAAAAGGGAGACACGAGAGUUGUCCCUUUCCGGCCAAAGGAUGUUGGGCUAAAGAGCUUUAAGCUGGCCCAGAAACGUCA_AAGAAACGUCAAAAC ((((..((((.(((((....).)))).))))..(.((((......)))).)...............))))......((((........))))...... ( -8.86 = -8.03 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:09 2006