
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,303,048 – 9,303,164 |
| Length | 116 |
| Max. P | 0.573456 |

| Location | 9,303,048 – 9,303,164 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.55 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -12.62 |
| Energy contribution | -13.71 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9303048 116 + 20766785 AGAUUCACUUACAGAAAAACAAAAAGUAC----ACAUGUGCAACAGGACACUAUGAGUUGUCUGUUAAUUGAAGGCUAUUUAUUUGCAAAAAGGAGCCUUACUCAUUAACUCAUCGAAAU ...(((...................((((----....))))....(((((........)))))((((((..((((((.(((........)))..))))))....)))))).....))).. ( -19.50) >DroSec_CAF1 187844 118 + 1 AGGUUCACUUCCAGAA-AACAAAACGUACAUACAUAUGUGGAACAGGACACUAUGAGGUGUCUGUUAAUUGAAGGCUAUUUAUUUGCAA-AAGAAGCCUUACUCAUUAACUCAUCGAAAU ...(((..((((....-.((.....))((((....))))))))..(((((((....)))))))((((((..((((((.(((........-))).))))))....)))))).....))).. ( -23.20) >DroSim_CAF1 173929 118 + 1 AGGUUCACUUCCAGAA-AACAAAAAGUACAUACAUAUGUGCAACAGGACACCAUGAGGUGUCUGUUAAUUGAAGGCUAUUUAUUUGCAA-AAGAAGCCUUACUCAUUAACUCAUCGAAAU .((.......))....-........((((((....))))))....(((((((....)))))))((((((..((((((.(((........-))).))))))....)))))).......... ( -28.00) >DroEre_CAF1 173519 92 + 1 AGGUUUACUUCCACAAAAACG---------------------------CACUGCGAAGUCUUUGUUACUUGAAGGCUAUUUGUUUGUAA-AAGGAGCCUUACUCGUUAACUCAUUGAAUU ((((((.(((..(((((..((---------------------------(...))).((((((((.....)))))))).....)))))..-)))))))))..................... ( -17.60) >DroYak_CAF1 172310 109 + 1 AGGUUUACUUCCACGAAAGUGAAGA----------AUGUGCAACAGGACACUGAAAAGUGUCUGUUAAUUGAAGGCUAUUUAUUUGUAA-AAGGAGCCUUACUCAUUAACUCAUUGAAUU ..((((((((((((....)))..))----------).))).))).(((((((....)))))))((((((..((((((.((((....)))-)...))))))....)))))).......... ( -30.30) >DroPer_CAF1 152881 91 + 1 AGA---ACUUCCAUUACAAUC----------------------CAAGCUAUAGAGAAACGCCAGUUAAGUGGAGGCUCUCCGACACGAG-AAUGAGCCUUUGACAUUGACUCCCCCA--- ...---...............----------------------...................((((((.(..((((((((((...)).)-)..))))))..)...))))))......--- ( -15.70) >consensus AGGUUCACUUCCAGAAAAACAAAAA__________AUGUGCAACAGGACACUAUGAAGUGUCUGUUAAUUGAAGGCUAUUUAUUUGCAA_AAGGAGCCUUACUCAUUAACUCAUCGAAAU .............................................(((((((....)))))))((((((..((((((.................))))))....)))))).......... (-12.62 = -13.71 + 1.09)

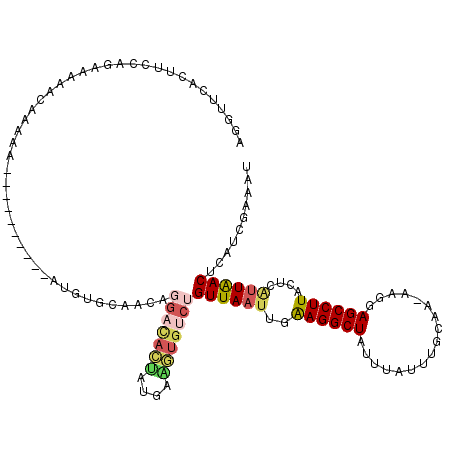
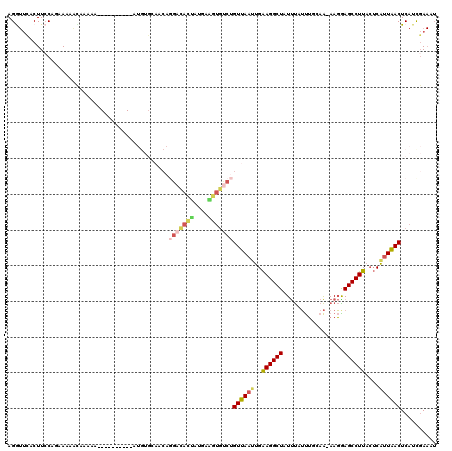
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:04 2006