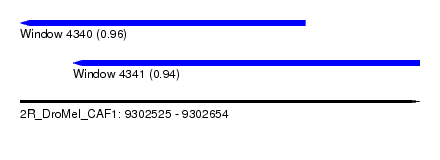
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,302,525 – 9,302,654 |
| Length | 129 |
| Max. P | 0.955973 |

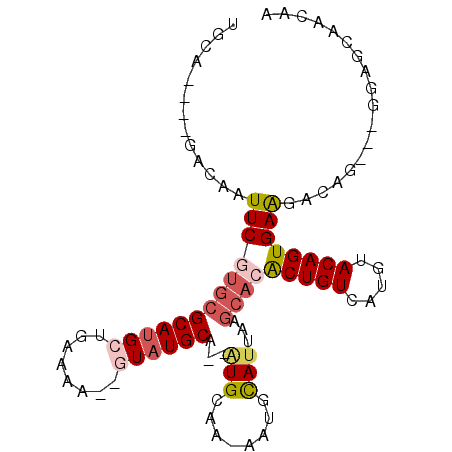
| Location | 9,302,525 – 9,302,617 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -11.27 |
| Energy contribution | -11.88 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9302525 92 - 20766785 UGCAAUUGGGAAAUUCGUGCGCAUGCUAAAAA--GUAUGCG--AUGCAA-AAUGCAUUAAGCAGACUGUCAUGUACAGUGAGGACUG---GGCGCGACAA ..............((.((((((((((....)--))))))(--((((..-...)))))..))))).((((.((..((((....))))---..)).)))). ( -28.20) >DroPse_CAF1 151923 87 - 1 ACCC-----CCAAUUCGUGGGCAUGCUGAAAA--AUAUGCA--GUGGAA-AAUGUAUUAAGCACGCUGUCAUGUACAGUGAAGACAG---CGAGCAGCGA ...(-----(((.....))))..((((.....--..(((((--.(....-).)))))...((.(((((((............)))))---)).)))))). ( -23.90) >DroYak_CAF1 171789 92 - 1 UGCAAUUGGGGAAUUCGUGCGCAUGCUGAAAA--GUAUGCG--AUGCAA-AAUGCAUUAAGCACACUGUCAUGUACAGUGAAGACUG---GGCGCGACAA ................(((((((((((....)--))))))(--((((..-...)))))..))))..((((.((..((((....))))---..)).)))). ( -33.90) >DroMoj_CAF1 279980 94 - 1 CGCAGU-UGACAAUUCGUGCGCAUGCCGAAAAGAGUAUGCAAAAUGCAAAAAUAUAUUAGGGCAGCUGUCGUCUACAGUGAGGACUG---GCA--AACAA .((..(-(((..(((..((((((((((.....).)))))).....)))..)))...)))).)).((((((.((......)).))).)---)).--..... ( -21.20) >DroAna_CAF1 178690 91 - 1 CGC----CGAGAAUUCGUGCGCAUGCUGAAAA--GUAUGCG--UUGCAA-AAUGCAUUAAGCACACUGUCAUGUACAGUGAAGACAGAAGGCGGCGACAA (((----((....((((((((((((((....)--)))))).--.(((..-...)))....))))(((((.....))))).......)))..))))).... ( -35.70) >DroPer_CAF1 152395 87 - 1 UGCC-----ACAAUUCGUGCGCAAGCUGAAAA--AUAUGCA--GUGGAA-AAUGUAUUAAGCACGCUGUCAUGUACAGUGAAGACAG---CGAGCAGCGA ((((-----((.....))).))).(((.....--..(((((--.(....-).)))))...((.(((((((............)))))---)).))))).. ( -25.20) >consensus UGCA____GACAAUUCGUGCGCAUGCUGAAAA__GUAUGCA__AUGCAA_AAUGCAUUAAGCACACUGUCAUGUACAGUGAAGACAG___GGAGCAACAA .............(((((((((((((........))))))...(((........)))...))))(((((.....)))))))).................. (-11.27 = -11.88 + 0.61)


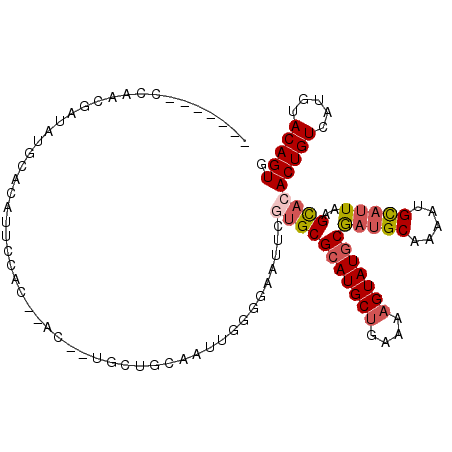
| Location | 9,302,542 – 9,302,654 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9302542 112 - 20766785 CACGCCUCCAACGAUAUGCACAUUCCACACACAGUGCUGCAAUUGGGAAAUUCGUGCGCAUGCUAAAAAGUAUGCGAUGCAAAAUGCAUUAAGCAGACUGUCAUGUACAGUG .(((..(((..((((.((((((((........)))).)))))))))))....)))(((((((((....)))))))(((((.....)))))..))..(((((.....))))). ( -29.50) >DroSim_CAF1 173429 103 - 1 -------ACAACGAUAUGCACAUUCCACACAC--UGCUGCAAUUGGGGAAUUCGUGCGCAUGCUGAAAAGUAUGCGAUGCAAAAUGCAUUAAGCAGACUGUCAUGUACAGUG -------..........(((((((((...((.--(......).)).)))))..))))(((((((....)))))))(((((.....)))))......(((((.....))))). ( -29.40) >DroEre_CAF1 173050 99 - 1 -------CCCACGA--UGCACAUUGCAC--AC--UGUUGCAGUUGGGGAAUUCGUGCGCAUGCUGAAAAGUAUGCGAUGCAAAAUGCAUUAAGUACACUGUCAUGUACAGUG -------((((...--((((((......--..--)).))))..))))......(((((((((((....)))))))(((((.....)))))..))))(((((.....))))). ( -34.80) >DroWil_CAF1 238805 89 - 1 ----------------UGCAGA---CACACAG--UAUUG-AAU-UGACAAUUCGUGCGGAUGCCGAAAAGUAUGCAAGGCAAAAUGUAUUAAGCUCACUGUCAUGUACAGUU ----------------((((((---((...((--(...(-(((-.....))))(((((..((((.............))))...)))))...)))...)))).))))..... ( -16.32) >DroYak_CAF1 171806 101 - 1 -------CCAACGAUAUGCACAUUGCAC--AC--UGCUGCAAUUGGGGAAUUCGUGCGCAUGCUGAAAAGUAUGCGAUGCAAAAUGCAUUAAGCACACUGUCAUGUACAGUG -------.(..((((.((((....((..--..--.))))))))))..).....(((((((((((....)))))))(((((.....)))))..))))(((((.....))))). ( -33.60) >DroAna_CAF1 178710 101 - 1 CACUCCCCCGCCCAUAUGCAUAUUUU-----C--UGGCGC----CGAGAAUUCGUGCGCAUGCUGAAAAGUAUGCGUUGCAAAAUGCAUUAAGCACACUGUCAUGUACAGUG .........((....(((((((((((-----(--.(....----)))))))..(((((((((((....)))))))).)))...))))))...)).((((((.....)))))) ( -30.80) >consensus _______CCAACGAUAUGCACAUUCCAC__AC__UGCUGCAAUUGGGGAAUUCGUGCGCAUGCUGAAAAGUAUGCGAUGCAAAAUGCAUUAAGCACACUGUCAUGUACAGUG .....................................................(((((((((((....)))))))(((((.....)))))..))))(((((.....))))). (-19.86 = -20.62 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:03 2006