| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,657,496 – 1,657,613 |
| Length | 117 |
| Max. P | 0.807757 |

| Location | 1,657,496 – 1,657,613 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -28.81 |
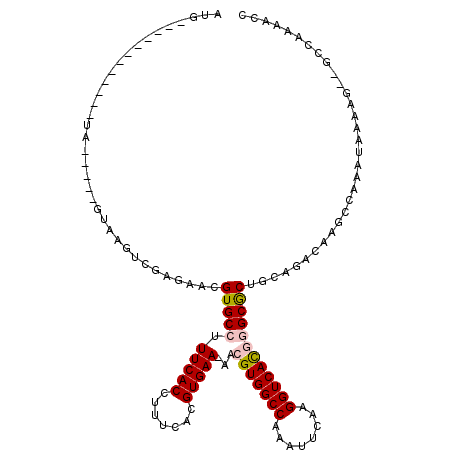
| Consensus MFE | -19.00 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.20 |
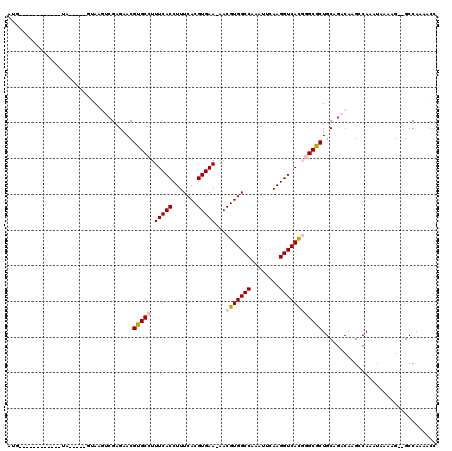
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
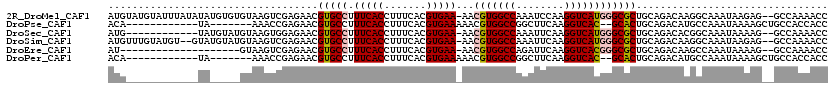
>2R_DroMel_CAF1 1657496 117 - 20766785 AUGUAUGUAUUUAUAUAUGUGUGUAAGUCGAGAACGUGCCUUUCACCUUUCACGUGAA-AACGUGGCCAAAUCCAAGGUCAUGGGCGCUGCAGACAAGGCAAAUAAGAG--GCCAAAACC .((((((....))))))(((.((((.((.....))(((((((((((.......)))))-).(((((((........)))))))))))))))).))).(((.........--)))...... ( -33.20) >DroPse_CAF1 73621 99 - 1 ACA------------UA-------AAACCGAGAACGUGCCUUUCACCUUUCACGUGAAAAACGUGGCCGGCUUCAAGGUCAC--GCACUGCAGACAUGCCAAAUAAAAGCUGCCACCACC ...------------..-------...........((((.((((((.......))))))...((((((........))))))--)))).((((.(.............)))))....... ( -22.52) >DroSec_CAF1 43353 105 - 1 AUG------------UAUGUAUGUAAGUGGAGAACGUGCCUUUCACCUUUCACGUGAA-AACGUGGCCAAAUUCAAGGUCAUGGGCGCUGCAGACACGGCAAAUAAAAG--GCCAAAACC ...------------..(((.((((.((.....))(((((((((((.......)))))-).(((((((........)))))))))))))))).))).(((.........--)))...... ( -30.40) >DroSim_CAF1 40224 115 - 1 AUGUUUGUAUGU--GUAUGUAUGUAAGUCGAGAACGUGCCUUUCACCUUUCACGUGAA-AACGUGGCCAAAUUCAAGGUCAUGGGCGCUGCAGACAAGGCAAAUAAGAG--GCCAAAACC .((((((((.((--((..((((((.........)))))).((((((.......)))))-).(((((((........))))))).)))))))))))).(((.........--)))...... ( -33.50) >DroEre_CAF1 47664 97 - 1 AU--------------------GUAAGUCGAGAACGUGCCUUUCACCUUUCACGUGAA-AACGUGGCCAGAUUCAAGGUCACGGGCGCUGCAGACAAGCCAAAUAAAAG--GCCAAAACC ..--------------------....(((..(...(((((((((((.......)))))-).(((((((........))))))))))))..).)))..(((........)--))....... ( -30.70) >DroPer_CAF1 67623 99 - 1 ACA------------UA-------AAACCGAGAACGUGCCUUUCACCUUUCACGUGAAAAACGUGGCCGGCUUCAAGGUCAC--GCACUGCAGACAUGCCAAAUAAAAGCUGCCACCACC ...------------..-------...........((((.((((((.......))))))...((((((........))))))--)))).((((.(.............)))))....... ( -22.52) >consensus AUG____________UA_____GUAAGUCGAGAACGUGCCUUUCACCUUUCACGUGAA_AACGUGGCCAAAUUCAAGGUCACGGGCGCUGCAGACAAGCCAAAUAAAAG__GCCAAAACC ...................................(((((.(((((.......)))))...(((((((........))))))))))))................................ (-19.00 = -19.20 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:57 2006