
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,285,217 – 9,285,318 |
| Length | 101 |
| Max. P | 0.837050 |

| Location | 9,285,217 – 9,285,318 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.59 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9285217 101 + 20766785 AAAUGUUGAUGGUCAAAAGUUGGC---UAACGCCUUGGCCAAAGCCAUGGCCAAAAAAGCAGCUCGAA-AUGCAGCCA---GCUAAAAGACAAAUAUCAAGUGAGUGG--- .....(((((((((...(((((((---(...(((.((((....)))).))).......(((.......-.))))))))---)))....)))...))))))........--- ( -31.30) >DroPse_CAF1 134934 106 + 1 AAAAGUUGAUGGUCAAAAGUUGGCGUCUGGUGAAGUGGCCAAAGCCAUGGCCAAAAAAGCUGCUCCAAGAGGCAGACA-C-GUUCAAAGGCAAACAGCAAACGAGAAG--- ....((((...((((.....))))((((..((((.((((((......)))))).....((((((......))))).).-.-.)))).))))...))))..........--- ( -29.70) >DroSec_CAF1 170222 101 + 1 AAAUGUUGAUGGUCAAAAGUUGGC---UAACGCCUUGGCCAAAGCCAUGGCCAAAAAAGCAGCUCGAA-AUGCAGCUA---GCUAAAAGACAAAUAUCAAGUGAGUUG--- .....(((((((((...(((((((---(...(((.((((....)))).))).......(((.......-.))))))))---)))....)))...))))))........--- ( -29.00) >DroSim_CAF1 156271 101 + 1 AAAUGUUGAUGGUCAAAAGUUGGC---UAACGCCUUGGCCAAAGCCAUGGCCAAAAAAGCAGCUCGAA-AUGCAGCCA---GCUAAAAGACAAAUAUCAAGUGAGUUA--- .....(((((((((...(((((((---(...(((.((((....)))).))).......(((.......-.))))))))---)))....)))...))))))........--- ( -31.30) >DroEre_CAF1 156540 99 + 1 --AUGUUGAUGGUCAAAAGUUGGC---UAACGCCUUGGCCAAAGCCAUGGCCAAAAAAGCAGCUCGAA-AGGCAGCCA---GCUAAAAGACAAAUAUCAAGCGAGUGG--- --...(((((((((...(((((((---(...((.(((((((......)))))))....)).((.(...-.))))))))---)))....)))...))))))........--- ( -33.30) >DroMoj_CAF1 255691 104 + 1 -AAAGUUUAUGGUCAAAAGUUAGC---AGGCA-CUUGGCCAAAGCCAUGGCCAAAAAUGCGUCUCACAGUUGCAGCCAACGGC-CAAAGACAAAUACCAA-CGAGCAGACC -...((((.(((((....((.((.---(.(((-.(((((((......)))))))...))).))).)).((((....)))))))-)).)))).........-.......... ( -28.80) >consensus AAAUGUUGAUGGUCAAAAGUUGGC___UAACGCCUUGGCCAAAGCCAUGGCCAAAAAAGCAGCUCGAA_AUGCAGCCA___GCUAAAAGACAAAUAUCAAGCGAGUAG___ .....((((((........(((((..........(((((((......)))))))....(((.........)))........)))))........))))))........... (-16.84 = -17.59 + 0.75)

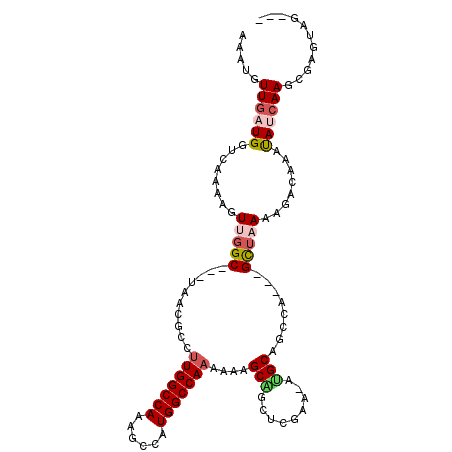
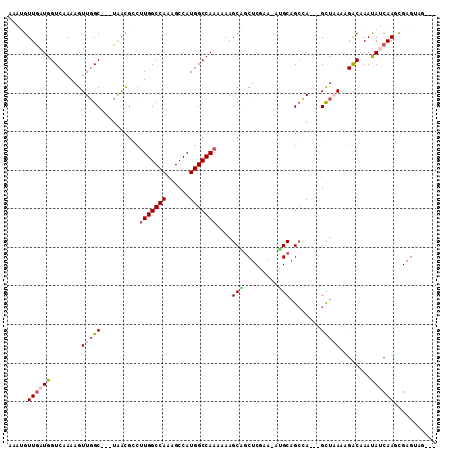
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:57 2006