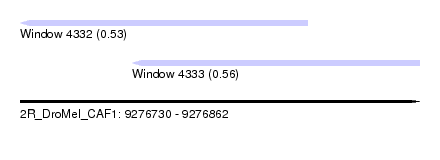
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,276,730 – 9,276,862 |
| Length | 132 |
| Max. P | 0.559052 |

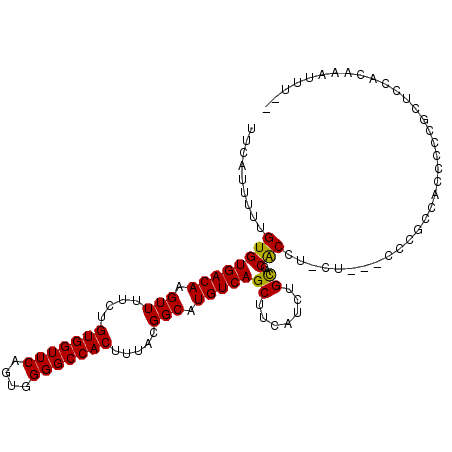
| Location | 9,276,730 – 9,276,825 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.99 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -14.07 |
| Energy contribution | -14.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.46 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9276730 95 - 20766785 GGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCAC-------------CACCCUCGCUCCAGAAUUUUUUUUUUUAUGGAAGCCAAGAGGUGGCGGCCUAUUUCACA ((((.......((..((.(((......))))))).(-------------((((((.(((((((((........))).))).)))..)).))))).))))......... ( -26.00) >DroSec_CAF1 161946 105 - 1 GGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCUACU---CCCGCCACACCCGCUCUACAAUUUUUUUUUUUAUGUUGGCCAAGGGGUGGCGGCCUAUUUCACA ((((.......((..((.(((......))))))).......---...(((((.(((......((((............)))).....))))))))))))......... ( -28.30) >DroSim_CAF1 147934 103 - 1 GGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCUACU---CCCGCCACACCCGCUCCACAAAUUU--UUUUUAUGUUGGCCAAGGGGUGGCGGCCUAUUUCACA ((((.......((..((.(((......))))))).......---...(((((.(((...(((((.....--......)).)))....))))))))))))......... ( -28.70) >DroEre_CAF1 148265 102 - 1 GGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCCCCUAGCCCCCCUGGCCCG--UCCACAAAUUU--UU--UAUGGGGGCCAAGGGGCGGCGGCCUAUUUCACA ((((.......((..((.(((......)))))))........(((((((((((((.--.(((.((....--.)--).)))))))).))))).)))))))......... ( -39.80) >DroYak_CAF1 146230 106 - 1 GGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCCGCUACCCCCACUACCACGGCUCCAAAAAUUU--UUUGUAUGGGGGCCAAGGGGCGGCGGCCUAUUUCACA ((((.......((...(((((......))).))...))((..((((.........((((((...((...--.)).....))))))..))))..))))))......... ( -31.50) >DroAna_CAF1 153712 92 - 1 GGCCACUUUACGGCGUGUCAGCUUCAUCUGUGGCGCCU------CCCGCUUC-CCUUCCCCAAAACUU-------UA--CGGGCCAAGGGGUGACGGCCUAUUUCACA ((((.......(((((..(((......)))..))))).------..(((..(-((((.(((.......-------..--.)))..))))))))..))))......... ( -29.90) >consensus GGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCU_CU___CCCGCCACCCCCGCUCCACAAAUUU__UUUUUAUGGGGGCCAAGGGGUGGCGGCCUAUUUCACA ((((.......((..((.(((......)))))))....................(((((((..........................))))))).))))......... (-14.07 = -14.57 + 0.50)

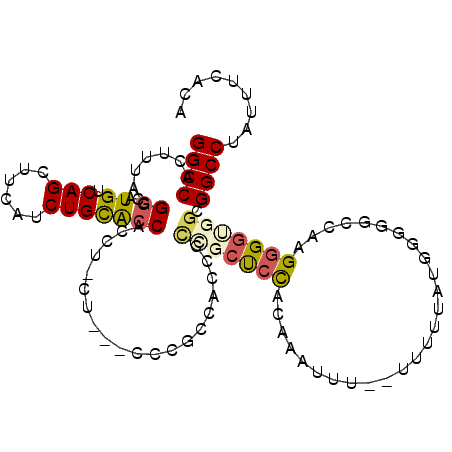
| Location | 9,276,767 – 9,276,862 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -18.71 |
| Energy contribution | -18.43 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9276767 95 - 20766785 UUCAUUUUUGUGUGACAAGUUUUCUGUGGUUCAGUGGGGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCAC-------------CACCCUCGCUCCAGAAUUUUUU .((((......)))).(((..(((((..((...((((.(((.......)))......((.......))....)-------------)))....))..)))))..))). ( -20.70) >DroSec_CAF1 161983 105 - 1 UUCAUUUUUGUGUGACAAGUUUUCUGUGGUUCAGUGGGGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCUACU---CCCGCCACACCCGCUCUACAAUUUUUU ..........((((...(((....((((((..(((((((((.......)))......((.......))....))))))---...))))))...))).))))....... ( -24.00) >DroSim_CAF1 147971 103 - 1 UUCAUUUUUGUGUGACAAGUUUUCUGUGGUUCAGUGGGGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCUACU---CCCGCCACACCCGCUCCACAAAUUU-- ......((((((.((...((....((((((..(((((((((.......)))......((.......))....))))))---...))))))...))))))))))...-- ( -26.90) >DroEre_CAF1 148300 104 - 1 UUCAUUUUUGUGUGACAAGUUUUCUGUGGUUCAGUGGGGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCCCCUAGCCCCCCUGGCCCG--UCCACAAAUUU-- ......((((((.(((.((....))..((..(((.(((((........((..((.(((......)))))))........))))).)))..)))--))))))))...-- ( -29.99) >DroYak_CAF1 146267 106 - 1 UUCAUUUUUGUGUGACAAGUUUUCUGUGGUUCAGUGGGGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCCGCUACCCCCACUACCACGGCUCCAAAAAUUU-- .((((......)))).((((((((((((((..(((((((........(((...(((((......))).))...)))....)))))))))))))).....)))))))-- ( -27.30) >DroAna_CAF1 153743 98 - 1 UUCAUUUUAGUGUGACAAGUUUUCUGUGGUUCACUGGGGCCACUUUACGGCGUGUCAGCUUCAUCUGUGGCGCCU------CCCGCUUC-CCUUCCCCAAAACUU--- .((((......)))).(((((((..(((((((....))))))).....(((((..(((......)))..))))).------........-........)))))))--- ( -24.50) >consensus UUCAUUUUUGUGUGACAAGUUUUCUGUGGUUCAGUGGGGCCACUUUACGGCAUGUCAGCUUCAUCUGCACCACCU_CU___CCCGCCACCCCCGCUCCACAAAUUU__ .........((((((((.(((....(((((((....))))))).....))).)))))((.......))..)))................................... (-18.71 = -18.43 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:55 2006