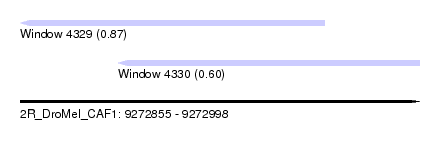
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,272,855 – 9,272,998 |
| Length | 143 |
| Max. P | 0.868319 |

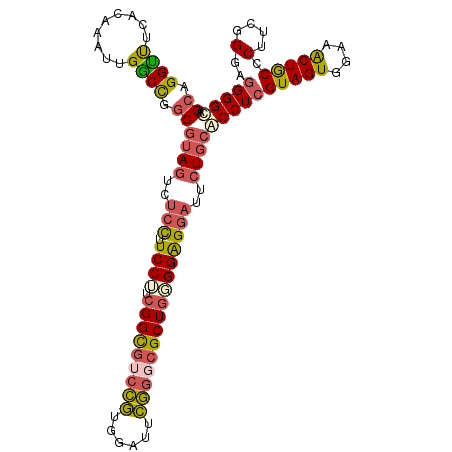
| Location | 9,272,855 – 9,272,964 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.77 |
| Mean single sequence MFE | -50.08 |
| Consensus MFE | -40.76 |
| Energy contribution | -41.13 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9272855 109 - 20766785 AGCAGGUUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGUGGAUUCGGGCGCUGUGGAGGAUUCUGCACCUCGUAGUGGAAACUGCCCUUCGGGAGGGGU .((.((((.........)))).))((((..((((((...(((((((((......))))))))).))))))..))))((((((((((....)))))((....)).))))) ( -52.40) >DroSec_CAF1 158083 109 - 1 AGCAGGCUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGUGGAUUCGGGCGCUGGGGAGGAUUCUGCACCUCGUAGUGGAAACUGCCCUUCGGGAGGGGU .((.((((.........)))).))((((..(((((((..(((((((((......))))))))))))))))..))))((((((((((....)))))((....)).))))) ( -57.40) >DroSim_CAF1 144055 109 - 1 AGCAGGUUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGCGGAUUCGGGCGCUGGGGAGGAUUCUGCACCUCGUAGUGGAAACUGCCCUUCGGGAGGGGU .((.((((.........)))).))((((..(((((((..(((((((((......))))))))))))))))..))))((((((((((....)))))((....)).))))) ( -53.90) >DroEre_CAF1 144479 109 - 1 AGCAGGUUGCACAAACUGGCCGGCGUAGUCUCCAUCCUUCGGUGUCCGUGGAUUCGGGCGCUGGGGAGGAUUCUGCACCUCGUAGUGGAAGCUACCCUUCGGAAGGGGC .((.((((.....)))).)).((.((((..(((.((((.(((((((((......))))))))))))))))..)))).))..(((((....))))).((((....)))). ( -45.80) >DroYak_CAF1 142285 109 - 1 AGCAGGCUUCACAAAUUGGCCGGCGUAGUCUCCUUCCGUCGGCGUCCGUGGACUCGGUCGCUGCGGAGGAUUCUGCACCUCGUAGUGGAAACUGCCCUUCGGAAGGGGC .((.((((.........)))).))((((..((((.(((.(((((.(((......))).))))))))))))..)))).(((((((((....))))).(....)..)))). ( -50.30) >DroMoj_CAF1 238436 109 - 1 AGCGGGUCUGAUGAAGGGACUAUCGUAGGCGAUUUCCUUGGGCUGCUUGGCAAUGAUGCGCUGCGGGGGAUUGUUGGCCUCGUAGUGGAAGCUGCCCUUCGGUAUGGGC .((..((((((...(((((..((((....)))).)))))((((.((((.(((....)))(((((((((.........)))))))))..)))).)))).)))).))..)) ( -40.70) >consensus AGCAGGUUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGUGGAUUCGGGCGCUGGGGAGGAUUCUGCACCUCGUAGUGGAAACUGCCCUUCGGGAGGGGC .((.((((.........)))).))((((..(((.((((.(((((((((......))))))))))))))))..))))((((((((((....))))).(....)..))))) (-40.76 = -41.13 + 0.37)

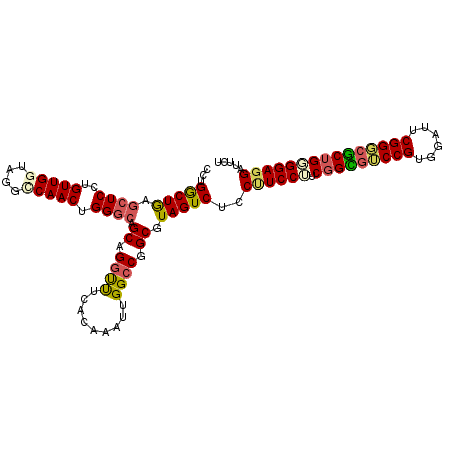
| Location | 9,272,890 – 9,272,998 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.16 |
| Mean single sequence MFE | -46.73 |
| Consensus MFE | -39.54 |
| Energy contribution | -39.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9272890 108 - 20766785 CCUGGCUAAGCUCCUGUUGAUAGGCCAACUGGGCAGCAGGUUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGUGGAUUCGGGCGCUGUGGAGGAUUCU .((((((((...(((((((.(((.....)))..))))))).........))))))))...((..((((((...(((((((((......))))))))).))))))..)) ( -46.40) >DroSec_CAF1 158118 108 - 1 CCUGGCUGAGCUCCUGUUGGUAGGCCAACUGGGCAGCAGGCUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGUGGAUUCGGGCGCUGGGGAGGAUUCU ...(((((.((.((.(((((....))))).)))).((.((((.........)))).)).)))))(((((((..(((((((((......)))))))))))))))).... ( -51.50) >DroSim_CAF1 144090 108 - 1 CCUGGCUGAGCUCCUGUUGGUAGGCCAACUGGGCAGCAGGUUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGCGGAUUCGGGCGCUGGGGAGGAUUCU .(((((..((((((((((((....))))).))).))).(......)...)..)))))...((..(((((((..(((((((((......))))))))))))))))..)) ( -49.60) >DroEre_CAF1 144514 108 - 1 CCUGGCUGAGCUCCUGUUGGAAGGUCAACUGGGCAGCAGGUUGCACAAACUGGCCGGCGUAGUCUCCAUCCUUCGGUGUCCGUGGAUUCGGGCGCUGGGGAGGAUUCU .((((((..(((((((((((....))))).))).))).((((.....))))))))))...((..(((.((((.(((((((((......))))))))))))))))..)) ( -44.40) >DroYak_CAF1 142320 108 - 1 CCUGGCUGAGCUCCUGUUGGUAGGUCAACUGGGCAGCAGGCUUCACAAAUUGGCCGGCGUAGUCUCCUUCCGUCGGCGUCCGUGGACUCGGUCGCUGCGGAGGAUUCU ((..(.(((.((((....)).)).))).)..))..((.((((.........)))).))..((..((((.(((.(((((.(((......))).))))))))))))..)) ( -45.30) >DroPer_CAF1 122435 108 - 1 CAUGACUGUAUUCCUGUUGAUAGGACAACUGGCCAGCAGGUCGCACAAAGUGGGCGGCGUAGUCCGCCUCCUGCUGUGGCCGAGGGCUCGGCAACUGCGGAGGACUUC ...(((((.....((((((.(((.....)))..))))))(((((.(.....).))))).)))))..(((((.((.((.(((((....))))).)).)))))))..... ( -43.20) >consensus CCUGGCUGAGCUCCUGUUGGUAGGCCAACUGGGCAGCAGGUUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGUGGAUUCGGGCGCUGGGGAGGAUUCU ...(((((.((((..(((((....))))).)))).((.((((.........)))).)).)))))..((((((.(((((((((......)))))))))))))))..... (-39.54 = -39.77 + 0.23)

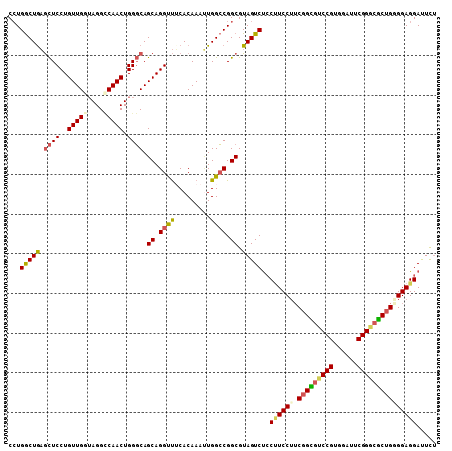
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:52 2006