
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,243,385 – 9,243,482 |
| Length | 97 |
| Max. P | 0.945433 |

| Location | 9,243,385 – 9,243,482 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -15.29 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
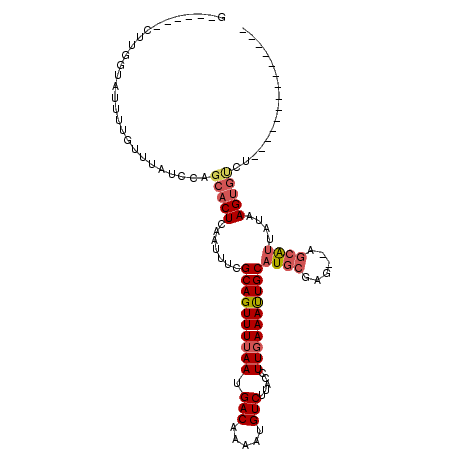
>2R_DroMel_CAF1 9243385 97 + 20766785 --------------AGACACUUAUAAUGCU--CCCGCAUGCAAUUUCAAGGUAAGACAGUUUGUCAUUAAAACUGCGAAAUUGAGUGCUAGAUAAACAAAAUACCAGGCAG--CC --------------.............(((--.((((((.(((((((.((.((((((.....))).)))...))..))))))).))))..(.....).........)).))--). ( -19.60) >DroVir_CAF1 186603 99 + 1 -----------CAUACUCUCUUAUAAUGCUCUAUCGCAUGCAAUUUGAAGGUAAGACAUUUUGUCAUUAAAACUGCGAAAUUGAGUGCUGUAUAAACAAAAUACGCUG-----UC -----------................((......((((.((((((..((.((((((.....))).)))...))...)))))).)))).((((.......))))))..-----.. ( -15.20) >DroPse_CAF1 102122 93 + 1 ----AGGA------AGGGACUC----CCGU--CCCGCAUGCAAUUUCAAGGUAAGACAGUUUGUCAUUAAAACUGCGAAAUUGAGUGCUGGAUAAACAAAAUACCAAAU------ ----.((.------.(((((..----..))--)))((((.(((((((.((.((((((.....))).)))...))..))))))).))))...............))....------ ( -27.50) >DroWil_CAF1 166569 96 + 1 ---------------AACACUUAUAAUGCUCUCUCGCAUGCAAUUUCAAGGUAAGACAUUUUGUCAUUAAAACUGCGAAAUUGAGUGCUAGAUAAACAAAAC--CACUCCA--AA ---------------................(((.((((.(((((((.((.((((((.....))).)))...))..))))))).)))).)))..........--.......--.. ( -18.50) >DroMoj_CAF1 207258 109 + 1 AGAGAGAAACACAUACACACUUAUAAUGCUAUCUCGCAUGCAGUUUCAAGGUAAGACAUUUUGUCAUUAAAACUGCGAAAUUGAGUGCUGUAUAAACAAAAUACGCAG------C ............(((((((((((..((((......))))(((((((.((.....(((.....))).)).))))))).....)))))).)))))...............------. ( -22.80) >DroAna_CAF1 127209 99 + 1 --------------AGACACUUAUAAUGCU--CCCGCAUGCAAUUUCAAGGUAAGACAGUUUGUCAUUAAAACUGCGAAAUUGAGUGCUAGAUAAACAAAAUGUAAAGGGGUCCC --------------.............((.--(((((((.(((((((.((.((((((.....))).)))...))..))))))).)))).......((.....))...)))))... ( -21.00) >consensus ______________AGACACUUAUAAUGCU__CCCGCAUGCAAUUUCAAGGUAAGACAGUUUGUCAUUAAAACUGCGAAAUUGAGUGCUAGAUAAACAAAAUACCAAG______C ...................................((((.(((((((.((.((((((.....))).)))...))..))))))).))))........................... (-15.29 = -15.32 + 0.03)


| Location | 9,243,385 – 9,243,482 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -14.72 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9243385 97 - 20766785 GG--CUGCCUGGUAUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCGGG--AGCAUUAUAAGUGUCU-------------- .(--((.((.((((.......))))..(((..((((((((((((((.......)))))))........)))))))..))))).--))).............-------------- ( -22.70) >DroVir_CAF1 186603 99 - 1 GA-----CAGCGUAUUUUGUUUAUACAGCACUCAAUUUCGCAGUUUUAAUGACAAAAUGUCUUACCUUCAAAUUGCAUGCGAUAGAGCAUUAUAAGAGAGUAUG----------- ..-----..((((((.......)))).))((((......((((((((((.(((.....)))))).....)))))))((((......)))).......))))...----------- ( -18.80) >DroPse_CAF1 102122 93 - 1 ------AUUUGGUAUUUUGUUUAUCCAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCGGG--ACGG----GAGUCCCU------UCCU---- ------...(((((.......)).)))(((..((((((((((((((.......)))))))........)))))))..)))(((--((..----..))))).------....---- ( -25.30) >DroWil_CAF1 166569 96 - 1 UU--UGGAGUG--GUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAAAUGUCUUACCUUGAAAUUGCAUGCGAGAGAGCAUUAUAAGUGUU--------------- ..--..(((((--.(...........).)))))......((((((((((.(((.....))).....))))))))))((((......))))..........--------------- ( -20.30) >DroMoj_CAF1 207258 109 - 1 G------CUGCGUAUUUUGUUUAUACAGCACUCAAUUUCGCAGUUUUAAUGACAAAAUGUCUUACCUUGAAACUGCAUGCGAGAUAGCAUUAUAAGUGUGUAUGUGUUUCUCUCU (------(((..((.......))..))))..........((((((((((.(((.....))).....))))))))))....((((.(((((((((....)))).)))))))))... ( -27.20) >DroAna_CAF1 127209 99 - 1 GGGACCCCUUUACAUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCGGG--AGCAUUAUAAGUGUCU-------------- ((....))...................(((((.......((((((((((.(((.....))).....))))))))))((((...--.))))....)))))..-------------- ( -21.80) >consensus G______CUUGGUAUUUUGUUUAUCCAGCACUCAAUUUCGCAGUUUUAAUGACAAAAUGUCUUACCUUGAAAUUGCAUGCGAG__AGCAUUAUAAGUGUCU______________ ...........................(((((.......((((((((((.(((.....))).....))))))))))((((......))))....)))))................ (-14.72 = -15.50 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:48 2006