| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,229,951 – 9,230,050 |
| Length | 99 |
| Max. P | 0.854399 |

| Location | 9,229,951 – 9,230,050 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -23.71 |
| Energy contribution | -24.88 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
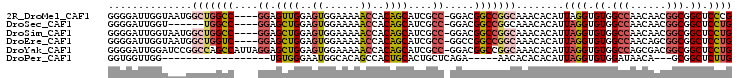
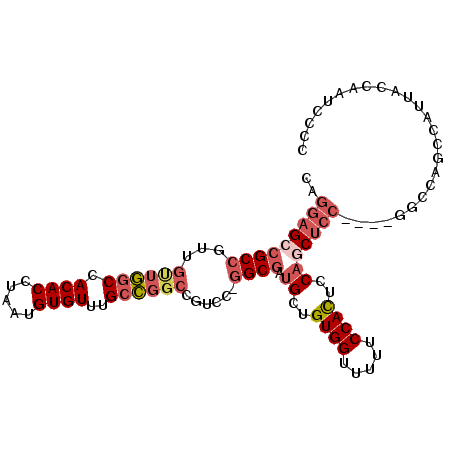
>2R_DroMel_CAF1 9229951 99 + 20766785 GGGGAUUGGUAAUGGCUGGCC----GGAGUUGGAGUGGAAAAACCACAGCAUCGCC-GGACGGCCGGCAAACACAUUAGGUGUGGCCAACAACGGCGGCUCCCG .((((.........((((.((----(..(((((.((((.....))))......(((-((....)))))...(((((...))))).)))))..))))))))))). ( -41.20) >DroSec_CAF1 123173 93 + 1 GGGGAUUGGU------UGGCC----GGAGCUGGAGUGGAAAAACCACAGCAUCGCC-GGACGGCCGGCAAACACAUUAGGUGUGGCCAACAACGGCGGCUCCUG ((((....((------(((((----(..((((..((......))..))))..)(((-((....)))))..((((.....)))))))))))........)))).. ( -39.40) >DroSim_CAF1 107872 99 + 1 GGGGAUUGGUAAUGGCUGGCC----GGAGCUGGAGUGGAAAAACCACAGCAUCGCC-GGACGGCCGGCAAACACAUUAGGUGUGGCCAACAACGGCGGCUCCUG .((((.(.((...(((((.((----((.((((..((......))..))))....))-)).)))))(((..((((.....)))).))).......)).).)))). ( -39.50) >DroEre_CAF1 108633 99 + 1 GGGGAUUGGUAAUGGCUGGUC----GGAGCUGGAGUGGAAAAACCACAGCAUCGCC-GGCCGGCCGGCAAACACAUUAGGUGUGGCCAACAGCGGCGGCUCCUG .((((.(.((...((((((((----((.((((..((......))..))))....))-))))))))(((..((((.....)))).))).......)).).)))). ( -43.90) >DroYak_CAF1 106456 103 + 1 GGGGAUUGGAUCCGGCCAGCCAUUAGGAGCUGGAGUGGAAAAACCACAGCAUCGCC-GGACGGCCGGCAAACACAUUAGGUGUGGCCAGCGACGGCGGCUCCUG ..((.((((......))))))..(((((((....((((.....)))).....((((-(....((.(((..((((.....)))).))).))..)))))))))))) ( -41.70) >DroPer_CAF1 89642 78 + 1 GGUGGUUGG------------------UGUGGGAAUGGCACAGCCACUGCACUGCUCAGA-----AACACACACAUUAGGUGUGGAUAACA---GCGGCUCUUG ((((((((.------------------(((.......)))))))))))...(((((....-----..(((((.......)))))......)---))))...... ( -24.20) >consensus GGGGAUUGGUAAUGGCUGGCC____GGAGCUGGAGUGGAAAAACCACAGCAUCGCC_GGACGGCCGGCAAACACAUUAGGUGUGGCCAACAACGGCGGCUCCUG ..............(((((((....((.((((..((......))..))))....)).....)))))))........((((.((.(((......))).)).)))) (-23.71 = -24.88 + 1.17)
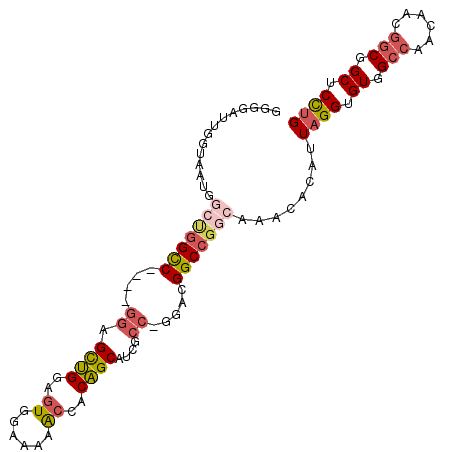

| Location | 9,229,951 – 9,230,050 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -21.64 |
| Energy contribution | -23.12 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
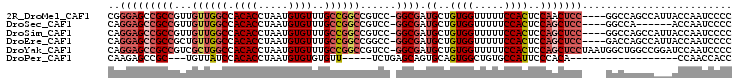
>2R_DroMel_CAF1 9229951 99 - 20766785 CGGGAGCCGCCGUUGUUGGCCACACCUAAUGUGUUUGCCGGCCGUCC-GGCGAUGCUGUGGUUUUUCCACUCCAACUCC----GGCCAGCCAUUACCAAUCCCC .((((......((.(((((((((((.....))))(((((((....))-)))))((..((((.....))))..)).....----))))))).))......)))). ( -37.40) >DroSec_CAF1 123173 93 - 1 CAGGAGCCGCCGUUGUUGGCCACACCUAAUGUGUUUGCCGGCCGUCC-GGCGAUGCUGUGGUUUUUCCACUCCAGCUCC----GGCCA------ACCAAUCCCC ..((((((((((..((((((.((((.....))))..))))))....)-)))).((..((((.....))))..)))))))----((...------.))....... ( -35.90) >DroSim_CAF1 107872 99 - 1 CAGGAGCCGCCGUUGUUGGCCACACCUAAUGUGUUUGCCGGCCGUCC-GGCGAUGCUGUGGUUUUUCCACUCCAGCUCC----GGCCAGCCAUUACCAAUCCCC ..((((((((((..((((((.((((.....))))..))))))....)-)))).((..((((.....))))..)))))))----((....))............. ( -37.20) >DroEre_CAF1 108633 99 - 1 CAGGAGCCGCCGCUGUUGGCCACACCUAAUGUGUUUGCCGGCCGGCC-GGCGAUGCUGUGGUUUUUCCACUCCAGCUCC----GACCAGCCAUUACCAAUCCCC ..((((((((((((((((((.((((.....))))..))))).))).)-)))).((..((((.....))))..)))))))----..................... ( -37.40) >DroYak_CAF1 106456 103 - 1 CAGGAGCCGCCGUCGCUGGCCACACCUAAUGUGUUUGCCGGCCGUCC-GGCGAUGCUGUGGUUUUUCCACUCCAGCUCCUAAUGGCUGGCCGGAUCCAAUCCCC .(((((((((((..((((((.((((.....))))..))))))....)-)))).((..((((.....))))..))))))))...((....))((((...)))).. ( -41.80) >DroPer_CAF1 89642 78 - 1 CAAGAGCCGC---UGUUAUCCACACCUAAUGUGUGUGUU-----UCUGAGCAGUGCAGUGGCUGUGCCAUUCCCACA------------------CCAACCACC ....((((((---(((....(((((.....)))))((((-----....))))..)))))))))(((.......))).------------------......... ( -21.30) >consensus CAGGAGCCGCCGUUGUUGGCCACACCUAAUGUGUUUGCCGGCCGUCC_GGCGAUGCUGUGGUUUUUCCACUCCAGCUCC____GGCCAGCCAUUACCAAUCCCC ..(((((((((...((((((.((((.....))))..))))))......)))).((..((((.....))))..)))))))......................... (-21.64 = -23.12 + 1.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:45 2006