
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,224,333 – 9,224,488 |
| Length | 155 |
| Max. P | 0.998682 |

| Location | 9,224,333 – 9,224,438 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.51 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -28.02 |
| Energy contribution | -28.27 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
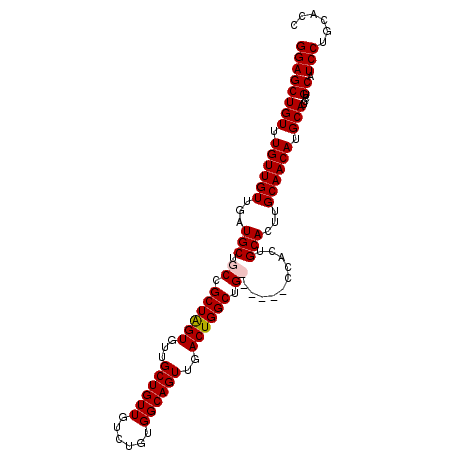
>2R_DroMel_CAF1 9224333 105 + 20766785 GGAGCUGUUUGUUGUUGAUGCUGCCGCUGGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUGCUAUUGCCACUGCACUUGCAACAUGCAUCCUGCAUCCUGCAUC ...((.(..(((....(((((((..((.(((((....((.((..(((((((((....))))))))).)).)).))))).))..)).)))))..)))..).))... ( -34.20) >DroSec_CAF1 117733 100 + 1 GGAGCUGUUUGUUGUUGAUGCUGCCGCUGGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUGCUAUUGCCACUGCACUUGCAACAUGCAUCCUGCAUCCU----- (((((.(..((((((((..((((((((.(..(.......)..).))))))))((..((((.......))))...))....))))).)))..).)).))).----- ( -33.20) >DroEre_CAF1 103181 98 + 1 GGAGCUGUUUGUUGUUGAUGC-GCCGCUAGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUG------CCACUGCACUUGCAACAUGCAUCCUGCAUCCUGCACC ((.((.(..(((....(((((-.......(((((((...(((..(((((((((....)))))------)))).)))...))))))))))))..)))..).)).)) ( -37.51) >DroYak_CAF1 100748 99 + 1 GGAGCUGUUUGUUGUUGAUGCUGCCGCUAGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUG------CCACUGCACUUGCAACAUGCAUUCUGCAUCCUGCACU ............(((.(((((....((..(((((((...(((..(((((((((....)))))------)))).)))...))))))))).....)))))..))).. ( -35.50) >consensus GGAGCUGUUUGUUGUUGAUGCUGCCGCUAGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUG______CCACUGCACUUGCAACAUGCAUCCUGCAUCCUGCACC ((((((((.((((((...(((.((.((((((...((((((......))))))..)))))).))..........)))...)))))).)))....)).)))...... (-28.02 = -28.27 + 0.25)

| Location | 9,224,333 – 9,224,438 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 91.51 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
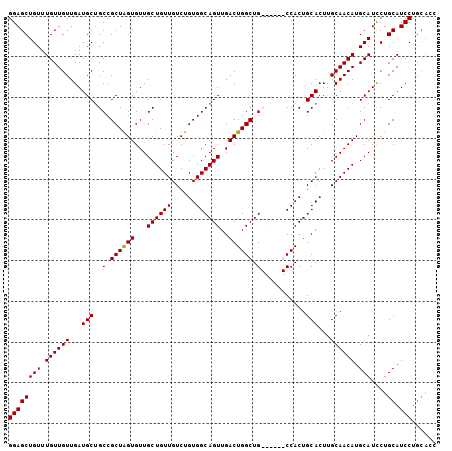
>2R_DroMel_CAF1 9224333 105 - 20766785 GAUGCAGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGGCAAUAGCAGCCAGUCAACUGCCACAGACAACAGCAACACCAGCGGCAGCAUCAACAACAAACAGCUCC (((((....(((.((.(((.(((((...((((((((((.......))).....)))).)))...))))))))...)).)))...)))))................ ( -29.20) >DroSec_CAF1 117733 100 - 1 -----AGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGGCAAUAGCAGCCAGUCAACUGCCACAGACAACAGCAACACCAGCGGCAGCAUCAACAACAAACAGCUCC -----..(((((....(((.((((((...(((.((((((...((.....))....)))))).).))...))))))...)))...)))))................ ( -26.90) >DroEre_CAF1 103181 98 - 1 GGUGCAGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGG------CAGCCAGUCAACUGCCACAGACAACAGCAACACUAGCGGC-GCAUCAACAACAAACAGCUCC (((((...((((.....))))(((((.((((..((((------(((........))))))).(....).....)))).)))))-)))))................ ( -32.00) >DroYak_CAF1 100748 99 - 1 AGUGCAGGAUGCAGAAUGCAUGUUGCAAGUGCAGUGG------CAGCCAGUCAACUGCCACAGACAACAGCAACACUAGCGGCAGCAUCAACAACAAACAGCUCC .((....(((((....(((.((((((...(((.((((------(((........))))))).).))...))))))...)))...)))))....)).......... ( -31.80) >consensus GGUGCAGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGG______CAGCCAGUCAACUGCCACAGACAACAGCAACACCAGCGGCAGCAUCAACAACAAACAGCUCC .......(((((....(((.((((((...(((.((((..........(((....))))))).).))...))))))...)))...)))))................ (-22.70 = -22.70 + -0.00)


| Location | 9,224,361 – 9,224,464 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.08 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -22.90 |
| Energy contribution | -26.77 |
| Covariance contribution | 3.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
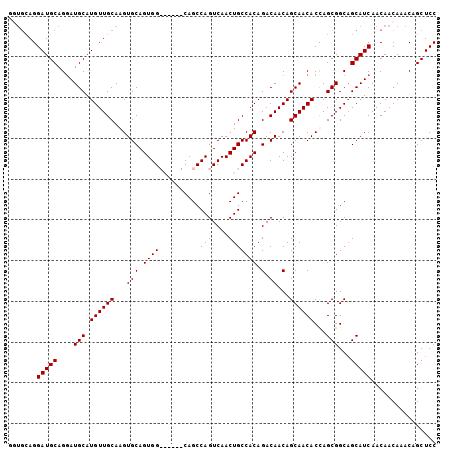
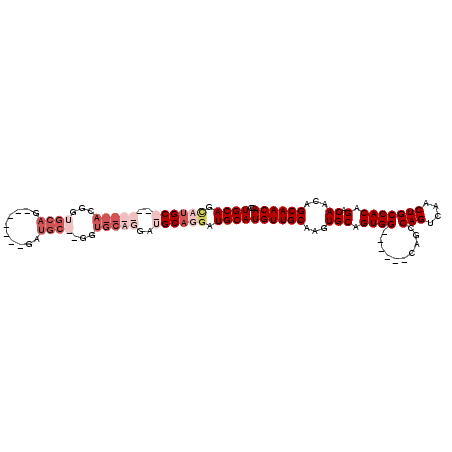
>2R_DroMel_CAF1 9224361 103 + 20766785 GGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUGCUAUUGCCACUGCACUUGCAACAUGCAUCCUGCAUCCUGCAUCCUGCAUC-------CUGCAGCGU-------GCAUGCUGCA (((((....((.((..(((((((((....))))))))).)).)).)))))((((.(((((((.(((((...(((.....)))..-------.))))).))-------))))).)))) ( -42.20) >DroSec_CAF1 117761 96 + 1 GGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUGCUAUUGCCACUGCACUUGCAACAUGCAUCCUGCAUCCU-------GCAUC-------CUGCACCGU-------GCAUGCUGCA .(((((((...(((..((((((((.(.(.....)).)))))))).)))...)))))))(((.(.(((((..(-------((...-------..)))..))-------))).).))). ( -36.40) >DroEre_CAF1 103208 97 + 1 AGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUG------CCACUGCACUUGCAACAUGCAUCCUGCAUCCUGCACC--------------CUGAGCCCUGCAACCUGCGAACUGCA .(((((((...(((..(((((((((....)))))------)))).)))...)))))))(((...((((...((((..--------------........))))...))))...))). ( -35.00) >DroYak_CAF1 100776 111 + 1 AGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUG------CCACUGCACUUGCAACAUGCAUUCUGCAUCCUGCACUCUGCAUCCUGCACUCUGCACCGUGCAACCUGCAACCUGCA .(((((((...(((..(((((((((....)))))------)))).)))...)))))))(((...((((...(((((..((((..........))))..)))))...))))...))). ( -43.80) >consensus AGUGUUGCUGUUGUCUGUGGCAGUUGACUGGCUG______CCACUGCACUUGCAACAUGCAUCCUGCAUCCUGCACC__GCAUC_______CUGCACCGU_______GCAAGCUGCA .(((((((...(((..(((((((((....)))))......)))).)))...)))))))(((.(.((((...(((((..((((..........))))..)))))...)))).).))). (-22.90 = -26.77 + 3.88)

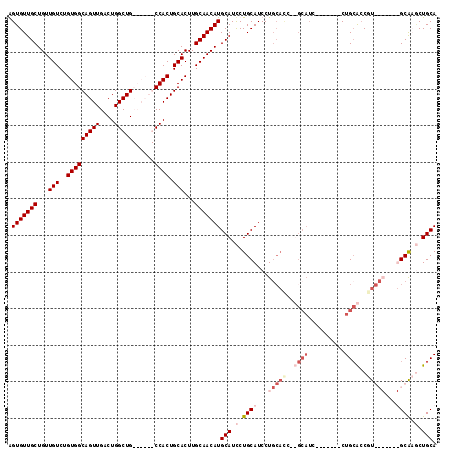
| Location | 9,224,361 – 9,224,464 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.08 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -19.25 |
| Energy contribution | -23.75 |
| Covariance contribution | 4.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
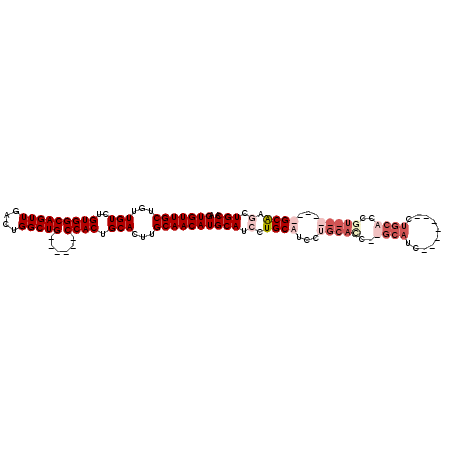
>2R_DroMel_CAF1 9224361 103 - 20766785 UGCAGCAUGC-------ACGCUGCAG-------GAUGCAGGAUGCAGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGGCAAUAGCAGCCAGUCAACUGCCACAGACAACAGCAACACC ((((((((((-------(..(((((.-------..(((.....)))...)))))..))))))))))).((((((((((.......))).....)))).)))................ ( -40.50) >DroSec_CAF1 117761 96 - 1 UGCAGCAUGC-------ACGGUGCAG-------GAUGC-------AGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGGCAAUAGCAGCCAGUCAACUGCCACAGACAACAGCAACACC (((....(((-------((..(((((-------.((((-------(..........))))).))))).)))))((((((...((.....))....))))))........)))..... ( -33.50) >DroEre_CAF1 103208 97 - 1 UGCAGUUCGCAGGUUGCAGGGCUCAG--------------GGUGCAGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGG------CAGCCAGUCAACUGCCACAGACAACAGCAACACU (((.....))).(((((...((.(..--------------..(((((.((((.....)))).))))).).)).((((------(((........)))))))........)))))... ( -35.80) >DroYak_CAF1 100776 111 - 1 UGCAGGUUGCAGGUUGCACGGUGCAGAGUGCAGGAUGCAGAGUGCAGGAUGCAGAAUGCAUGUUGCAAGUGCAGUGG------CAGCCAGUCAACUGCCACAGACAACAGCAACACU ((((..(((((..((((((..((((..........))))..))))))..)))))..))))((((((...(((.((((------(((........))))))).).))...)))))).. ( -47.80) >consensus UGCAGCAUGC_______ACGGUGCAG_______GAUGC__GGUGCAGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGG______CAGCCAGUCAACUGCCACAGACAACAGCAACACC ((((.((((((..(((((...((((..........))))...)))))..)))))).))))((((((...(((.((((..........(((....))))))).).))...)))))).. (-19.25 = -23.75 + 4.50)

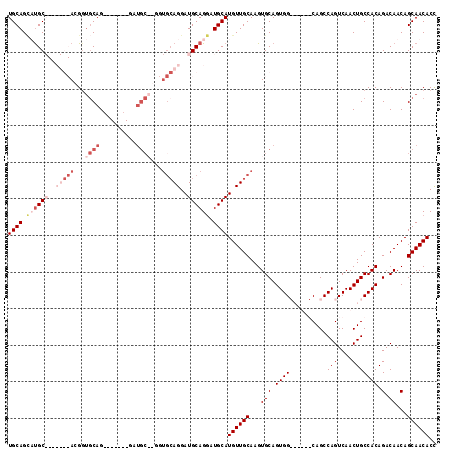
| Location | 9,224,398 – 9,224,488 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -14.08 |
| Energy contribution | -16.12 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9224398 90 + 20766785 UUGCCACUGCACUUGCAACAUGCAUCCUGCAUCCUGCAUCCUGCAUC-------CUGCAGCGU-------GCAUGCUGCAACUAGCAACUCGACGCGUGUCUGG ...(((..(((((((((.(((((((.(((((...(((.....)))..-------.))))).))-------))))).)))))...((........)))))).))) ( -33.40) >DroSec_CAF1 117798 83 + 1 UUGCCACUGCACUUGCAACAUGCAUCCUGCAUCCU-------GCAUC-------CUGCACCGU-------GCAUGCUGCAACUAGCAACUCGACGCGUGUCUGG ...(((..(((((((((.(((((((..((((....-------.....-------.))))..))-------))))).)))))...((........)))))).))) ( -25.40) >DroSim_CAF1 102004 83 + 1 UUGCCACUGCACUUGCAACAUGCAUCCUGCAUCCU-------GCAUC-------CUGCACCGU-------GCAUGCUGCAACUAGCAACUCGACGCGUGUCUGG ...(((..(((((((((.(((((((..((((....-------.....-------.))))..))-------))))).)))))...((........)))))).))) ( -25.40) >DroEre_CAF1 103242 87 + 1 ---CCACUGCACUUGCAACAUGCAUCCUGCAUCCUGCACC--------------CUGAGCCCUGCAACCUGCGAACUGCAACUCGCAACUCGACGCGUGUCUGG ---(((.(((....)))((((((.((.((((....((...--------------....))..))))...(((((........)))))....)).)))))).))) ( -20.10) >DroYak_CAF1 100810 101 + 1 ---CCACUGCACUUGCAACAUGCAUUCUGCAUCCUGCACUCUGCAUCCUGCACUCUGCACCGUGCAACCUGCAACCUGCAACGAGCAACUCGACGCGUGUCUGG ---(((.(((....)))((((((....((((...((((...(((((..(((.....)))..)))))...))))...)))).((((...))))..)))))).))) ( -30.60) >consensus UUGCCACUGCACUUGCAACAUGCAUCCUGCAUCCUGCA____GCAUC_______CUGCACCGU_______GCAUGCUGCAACUAGCAACUCGACGCGUGUCUGG ...(((..(((((((((.(((((.....((.....)).....(((..........)))............))))).)))))...((........)))))).))) (-14.08 = -16.12 + 2.04)

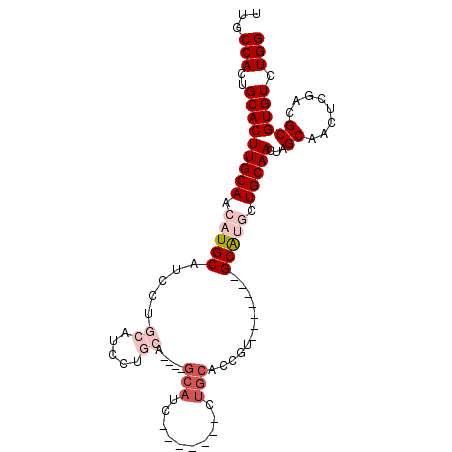
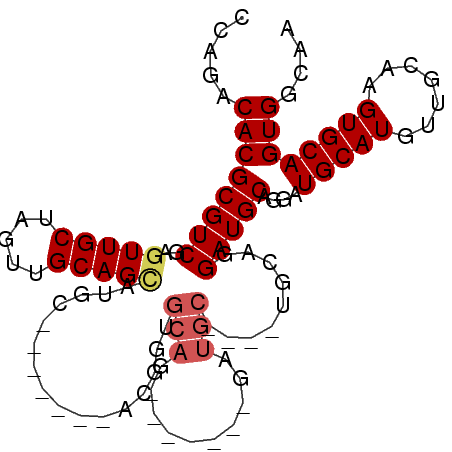
| Location | 9,224,398 – 9,224,488 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.54 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9224398 90 - 20766785 CCAGACACGCGUCGAGUUGCUAGUUGCAGCAUGC-------ACGCUGCAG-------GAUGCAGGAUGCAGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGGCAA ...(((....)))..((..(((.(((((((((((-------(..(((((.-------..(((.....)))...)))))..)))))))))))).)..))..)).. ( -40.50) >DroSec_CAF1 117798 83 - 1 CCAGACACGCGUCGAGUUGCUAGUUGCAGCAUGC-------ACGGUGCAG-------GAUGC-------AGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGGCAA ...(((....)))..(((((.....))))).(((-------.((.((((.-------..(((-------((.((((.....)))).)))))..)))).))))). ( -32.00) >DroSim_CAF1 102004 83 - 1 CCAGACACGCGUCGAGUUGCUAGUUGCAGCAUGC-------ACGGUGCAG-------GAUGC-------AGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGGCAA ...(((....)))..(((((.....))))).(((-------.((.((((.-------..(((-------((.((((.....)))).)))))..)))).))))). ( -32.00) >DroEre_CAF1 103242 87 - 1 CCAGACACGCGUCGAGUUGCGAGUUGCAGUUCGCAGGUUGCAGGGCUCAG--------------GGUGCAGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGG--- .....((((((.((((((((((.((((.....)))).)))))..))))..--------------..(((((.((((.....)))).))))).)))).))).--- ( -33.20) >DroYak_CAF1 100810 101 - 1 CCAGACACGCGUCGAGUUGCUCGUUGCAGGUUGCAGGUUGCACGGUGCAGAGUGCAGGAUGCAGAGUGCAGGAUGCAGAAUGCAUGUUGCAAGUGCAGUGG--- .....(((((((((((...))).(((((..(((((..((((((........))))))..)))))..))))))))))....(((((.......)))))))).--- ( -38.70) >consensus CCAGACACGCGUCGAGUUGCUAGUUGCAGCAUGC_______ACGGUGCAG_______GAUGC____UGCAGGAUGCAGGAUGCAUGUUGCAAGUGCAGUGGCAA .....((((((((..(((((.....)))))................(((..........))).........)))))....(((((.......)))))))).... (-18.86 = -19.54 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:42 2006