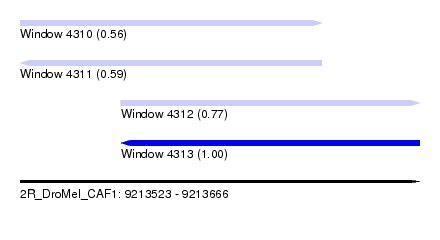
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,213,523 – 9,213,666 |
| Length | 143 |
| Max. P | 0.995028 |

| Location | 9,213,523 – 9,213,631 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9213523 108 + 20766785 UUUGUCGAGCACCUACUUUUCGGUGCUGCUCGGCUCUAGCACUCACCUUUCCGGUAAUUUGUGCAU-UUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUU- ...((((((((.(.(((....)))).))))))))....((....(((.....)))...((((((..-....))))))......))........((((.....))))...- ( -28.90) >DroSec_CAF1 107146 108 + 1 UUUGUCGAGCACCUACUUUUCGGUGCUGCUCGGCUCCAGCACUCACCUUGCCGGUAAUUUGUGCAU-UUUCGCAUAAUAUUUAGCACAAUUUUCCGCAUUUUGCGGCUU- ...((((((((.(.(((....)))).))))))))...............(((((.((.((((((..-................)))))).)).))((.....)))))..- ( -29.47) >DroSim_CAF1 90185 108 + 1 UUUGUCGAGCACCUACUUUUCGGUGCUGCUCGGCUCCAGCACUCACCUUGCCGGUAAUUUGUGCAU-UUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUU- ...((((((((.(.(((....)))).))))))))...............(((((.((.((((((..-................)))))).)).))((.....)))))..- ( -29.47) >DroEre_CAF1 93035 108 + 1 UUUGUCGAGCACCUACUUUUCGGUGCUACUCGGCGCCAGCACUCACCUUGCCGGUAAUUUGUGCAU-UUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUU- ...((((((((((........)))))...)))))(((.(((.......(((.((.((.((((((..-................)))))).)).)))))...))))))..- ( -31.77) >DroYak_CAF1 89465 108 + 1 UUUGUCGAGCACCUACUUUUCGGUGCUACUCGGCUCCAGCACUCACCUUGCCGGUAAUUUGUGCAU-UUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUU- ...((((((((((........)))))...)))))...............(((((.((.((((((..-................)))))).)).))((.....)))))..- ( -28.97) >DroPer_CAF1 76913 90 + 1 UUUCUCCAG------GCUUUCGACGCUCCUCGA------CACUCACCUUGAAUGUAAUUUGUGCACAUUUCUCGACAGAUU--------UUUGCCACAUUUUGUGGCCUG ......(((------((..((((......))))------....(((...((((((.....(.(((.(..(((....)))..--------).)))))))))).)))))))) ( -17.80) >consensus UUUGUCGAGCACCUACUUUUCGGUGCUGCUCGGCUCCAGCACUCACCUUGCCGGUAAUUUGUGCAU_UUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUU_ ...((((((((((........))))))...))))....((((..(((.....))).....)))).............................((((.....)))).... (-19.88 = -20.80 + 0.92)

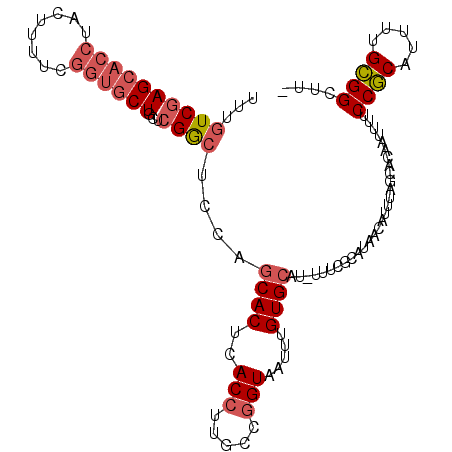
| Location | 9,213,523 – 9,213,631 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -30.31 |
| Consensus MFE | -21.75 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9213523 108 - 20766785 -AAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAA-AUGCACAAAUUACCGGAAAGGUGAGUGCUAGAGCCGAGCAGCACCGAAAAGUAGGUGCUCGACAAA -...((((.....))))....((((......((((..((....-..((((...(((((.....)))))))))....))..))))(((((........)))))...)))). ( -31.30) >DroSec_CAF1 107146 108 - 1 -AAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUAUUAUGCGAAA-AUGCACAAAUUACCGGCAAGGUGAGUGCUGGAGCCGAGCAGCACCGAAAAGUAGGUGCUCGACAAA -...((((.....))))....((((((................-..))))))....((((((.......)))))).(.((((((.(((......)).).)))))).)... ( -32.07) >DroSim_CAF1 90185 108 - 1 -AAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAA-AUGCACAAAUUACCGGCAAGGUGAGUGCUGGAGCCGAGCAGCACCGAAAAGUAGGUGCUCGACAAA -...((((.....))))....((((((....((.....))...-..))))))....((((((.......)))))).(.((((((.(((......)).).)))))).)... ( -32.30) >DroEre_CAF1 93035 108 - 1 -AAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAA-AUGCACAAAUUACCGGCAAGGUGAGUGCUGGCGCCGAGUAGCACCGAAAAGUAGGUGCUCGACAAA -...((((.....))))....((((((....((.....))...-..))))))..(((((((..(((....)))...)))).)))(((((........)))))........ ( -32.40) >DroYak_CAF1 89465 108 - 1 -AAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAA-AUGCACAAAUUACCGGCAAGGUGAGUGCUGGAGCCGAGUAGCACCGAAAAGUAGGUGCUCGACAAA -...((((.....))))....((((((....((.....))...-..))))))..(((((((..(((....)))...)))).)))(((((........)))))........ ( -32.40) >DroPer_CAF1 76913 90 - 1 CAGGCCACAAAAUGUGGCAAA--------AAUCUGUCGAGAAAUGUGCACAAAUUACAUUCAAGGUGAGUG------UCGAGGAGCGUCGAAAGC------CUGGAGAAA (((((((((.....(((((..--------....))))).....)))).........(((((.....)))))------((((......))))..))------)))...... ( -21.40) >consensus _AAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAA_AUGCACAAAUUACCGGCAAGGUGAGUGCUGGAGCCGAGCAGCACCGAAAAGUAGGUGCUCGACAAA ....((((.....))))..............((((...........((((...(((((.....)))))))))...........((((((........)))))).)))).. (-21.75 = -22.58 + 0.84)

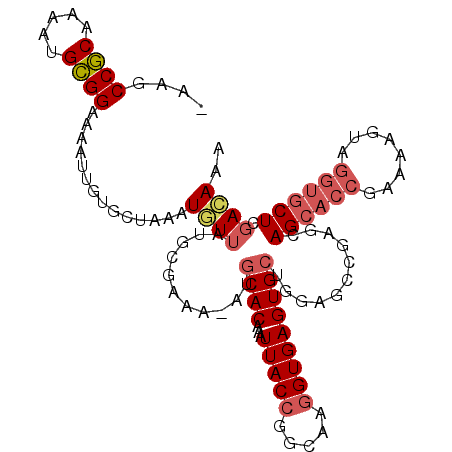
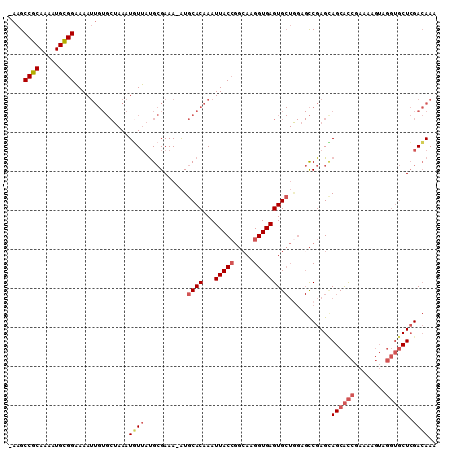
| Location | 9,213,559 – 9,213,666 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -32.76 |
| Consensus MFE | -27.03 |
| Energy contribution | -27.27 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9213559 107 + 20766785 UAGCACUCACCUUUCCGGUAAUUUGUGCAUUUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUUCGAGUGACAGAGUGAGGAAUGCUGUGGGAUGCUGC ((((((((((..((((......((((((......))))))(((((.((((......((((.....))))......))).).))))).))))....))))).))))). ( -33.30) >DroSec_CAF1 107182 107 + 1 CAGCACUCACCUUGCCGGUAAUUUGUGCAUUUUCGCAUAAUAUUUAGCACAAUUUUCCGCAUUUUGCGGCUUCGAGUGACAGAGUGAGGAAUGCUGUGGGAUGCUGA ((((((((((...((.(.(((.((((((......))))))...))).)......((((.(((((((..((.....))..))))))).)))).)).))))).))))). ( -34.60) >DroSim_CAF1 90221 107 + 1 CAGCACUCACCUUGCCGGUAAUUUGUGCAUUUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUUCGAGUGACAGAGUGAGGAAUGCUGUGGGAUGCUGC ((((((((((...((.(.(((.((((((......))))))...))).)......((((.(((((((..((.....))..))))))).)))).)).))))).))))). ( -33.20) >DroEre_CAF1 93071 107 + 1 CAGCACUCACCUUGCCGGUAAUUUGUGCAUUUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUUCGAGUGACAGAGCCAGGAAUCCUGCGGAAUGCUGG (((((.......(((..(....((((((......))))))...)..))).....(((((((..((.(((((..(.....)..)))).).))...)))))))))))). ( -31.80) >DroYak_CAF1 89501 107 + 1 CAGCACUCACCUUGCCGGUAAUUUGUGCAUUUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUUCGAGUGACAGAGCGAAGAAUGCUGUGGCAUGCUGC (((((.......(((((.....((((((......))))))....(((((.......((((.....))))(((((..(....)..)))))..))))))))))))))). ( -30.91) >consensus CAGCACUCACCUUGCCGGUAAUUUGUGCAUUUUCGCAUAACAUUUAGCACAAUUUUCCGCAUUUUGCGGCUUCGAGUGACAGAGUGAGGAAUGCUGUGGGAUGCUGC ((((((((((......((.((.((((((..................)))))).)).))(((((((...(((..(.....)..)))..))))))).))))).))))). (-27.03 = -27.27 + 0.24)



| Location | 9,213,559 – 9,213,666 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -27.95 |
| Energy contribution | -28.35 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9213559 107 - 20766785 GCAGCAUCCCACAGCAUUCCUCACUCUGUCACUCGAAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAAAUGCACAAAUUACCGGAAAGGUGAGUGCUA ............(((((((.((..((((..........((((.....))))....((((((....((.....)).....)))))).....))))..)).))))))). ( -26.20) >DroSec_CAF1 107182 107 - 1 UCAGCAUCCCACAGCAUUCCUCACUCUGUCACUCGAAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUAUUAUGCGAAAAUGCACAAAUUACCGGCAAGGUGAGUGCUG ...........(((((...(((((..((((........((((.....))))....((((((..................))))))......))))..)))))))))) ( -32.37) >DroSim_CAF1 90221 107 - 1 GCAGCAUCCCACAGCAUUCCUCACUCUGUCACUCGAAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAAAUGCACAAAUUACCGGCAAGGUGAGUGCUG ...........(((((...(((((..((((........((((.....))))....((((((....((.....)).....))))))......))))..)))))))))) ( -32.60) >DroEre_CAF1 93071 107 - 1 CCAGCAUUCCGCAGGAUUCCUGGCUCUGUCACUCGAAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAAAUGCACAAAUUACCGGCAAGGUGAGUGCUG .((((((((..((((...))))((..((((........((((.....))))....((((((....((.....)).....))))))......))))..)))))))))) ( -34.30) >DroYak_CAF1 89501 107 - 1 GCAGCAUGCCACAGCAUUCUUCGCUCUGUCACUCGAAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAAAUGCACAAAUUACCGGCAAGGUGAGUGCUG .((((((((....)).....((((..((((........((((.....))))....((((((....((.....)).....))))))......))))..)))))))))) ( -31.80) >consensus GCAGCAUCCCACAGCAUUCCUCACUCUGUCACUCGAAGCCGCAAAAUGCGGAAAAUUGUGCUAAAUGUUAUGCGAAAAUGCACAAAUUACCGGCAAGGUGAGUGCUG ...........(((((...(((((..((((........((((.....))))....((((((..................))))))......))))..)))))))))) (-27.95 = -28.35 + 0.40)

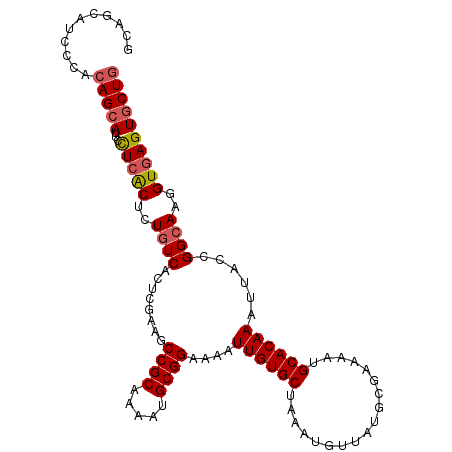
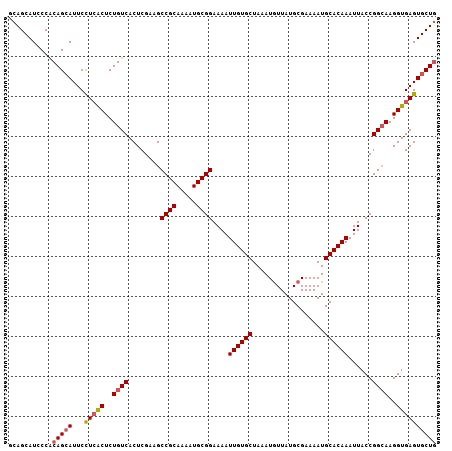
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:37 2006