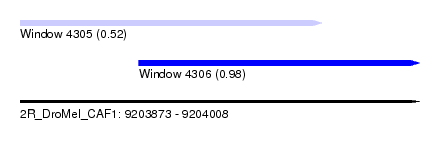
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,203,873 – 9,204,008 |
| Length | 135 |
| Max. P | 0.976527 |

| Location | 9,203,873 – 9,203,975 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -16.68 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9203873 102 + 20766785 CACUUCAGCCGACGCUUAAGUUGCACAAAAUGUAAUACAUUUGAUUGCACUCAUGUUCAAGUU---CUGUGGCCAAAGUGGCUGCCGGAGCUGGAG--------AGCUGGAGU .(((((((((..(((...(((.(((..(((((.....)))))...))))))...))...((((---(((..(((.....)))..).)))))))..)--------.)))))))) ( -35.10) >DroPse_CAF1 70241 102 + 1 CACUUCAGCCGGCGCUUAAGUUGCACAAAAUGUAAUACAUUUGAUUGCACUCAUGUUCAAGUU---CUGUUGCCAAAGUGGCUGCCUCUGCCGUCUCU--------CUCGCUC ......(((.(((((..((.(((.(((...((((((.(....)))))))....))).))).))---..).))))..((((((.......)))).))..--------...))). ( -20.50) >DroSec_CAF1 97279 100 + 1 CACUUCAGCCGACGCUUAAGUUGCACAAAAUGUAAUACAUUUGAUUGCACUCAUGUUCAAGUU---CUGUGGCCAAAGUGGUUGCCGGAGCUGG----------AGCUAGAGU .((((.(((....))).)))).(((..(((((.....)))))...)))((((..((((.((((---(((..(((.....)))..).)))))).)----------)))..)))) ( -32.80) >DroWil_CAF1 112872 89 + 1 CACUUCAGCCGCCGCUUAAGUUGCACAAAAUGUAAUACAUUUGAUUGCACUCAUGUUCAAGUUGUGCUGCUGCCAAAGUGGCUGCCGAC------------------------ ....((.((.(((((((.(((.((((((..((....((((..(......)..)))).))..)))))).)))....))))))).)).)).------------------------ ( -27.30) >DroAna_CAF1 87181 107 + 1 CACUUCAGCCGACGCUUAAGUUGCACAAAAUGUAAUACAUUUGAUUGCACUCAUGCUCAAGUU---CUGUGGCCAAAGUGGCUGCUACUGCUGCUGCUUUUGCU---UUCUGC (((...(((.((.((...(((.(((..(((((.....)))))...))))))...))))..)))---..)))((.((((..((.((....)).))..)))).)).---...... ( -27.60) >DroPer_CAF1 69649 106 + 1 CACUUCAGCCGGCGCUUAAUUAGCACAAAAUGUAAUACAUUUGAUUGCACUCAUGUUCAAGUU---CUGUUGCCAAAGUGGCGGCCUCUGCCGUCUCUCU----CUCUCGCAC .......((.((((((.....))).........((((.((((((..(((....))))))))).---.)))))))..((.((((((....)))))).))..----.....)).. ( -23.90) >consensus CACUUCAGCCGACGCUUAAGUUGCACAAAAUGUAAUACAUUUGAUUGCACUCAUGUUCAAGUU___CUGUGGCCAAAGUGGCUGCCGCUGCUGG____________CUCGAGC .((((.(((....))).)))).(((..(((((.....)))))...)))....................((((((.....))))))............................ (-16.68 = -16.73 + 0.06)


| Location | 9,203,913 – 9,204,008 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.33 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -17.64 |
| Energy contribution | -18.17 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9203913 95 + 20766785 UUGAUUGCACUCAUGUUCAAGUUCUGUGGCCAAAGUGGCUGCCGGAGCUGGAG----------AGCUGGAGU-----CUCAAAGGAGGAGGAGAGGAAAA-AAAAGUGCAG------ .....((((((((.((((.(((((((..(((.....)))..).))))))...)----------)))))...(-----(((....))))............-...)))))).------ ( -29.60) >DroPse_CAF1 70281 99 + 1 UUGAUUGCACUCAUGUUCAAGUUCUGUUGCCAAAGUGGCUGCCUCUGCCGUCUCU----------CUCGCUC-----UCCAAAAAACGAAGGGG--AAAA-AAGAGUAAAGUGAAAC .......((((..............((.(((.....))).))((((....((((.----------.(((...-----.........)))..)))--)...-.))))...)))).... ( -20.10) >DroSec_CAF1 97319 91 + 1 UUGAUUGCACUCAUGUUCAAGUUCUGUGGCCAAAGUGGUUGCCGGAGCUGG------------AGCUAGAGU-----CUCAAAGGAGGAGGAGC--AAAA-AAAAGUGCAG------ .....((((((...((((.(((((((..(((.....)))..).)))))).)------------)))..(..(-----(((.......))))..)--....-...)))))).------ ( -30.40) >DroSim_CAF1 80702 92 + 1 UUGAUUGCACUCAUGUUCAAGUUCUGUGGCCAAAGUGGCUGCCGGAGCUGG------------AGCUAGAGU-----CUCAAAGGAGGAGGAGC--AAAAAAAAAGUGCAG------ .....((((((...((((.(((((((..(((.....)))..).)))))).)------------)))..(..(-----(((.......))))..)--........)))))).------ ( -31.80) >DroEre_CAF1 83805 102 + 1 UUGAUUGCACUCAUGUUCAAGUUCUGUGGCCAAAGUGGCUGCCGGAGUUGGAGCAGGAGCUGGAGCUGGAGCUGCUCCUCAAAGGAGGAGGA------AA---AAGCGCAG------ .....(((.(((.(((((((.(((.(..(((.....)))..).))).)).))))).))).....(((.....(.((((((....)))))).)------..---.)))))).------ ( -37.50) >DroYak_CAF1 79635 99 + 1 UUGAUUGCACUCAUGUUCAAGUUCUGUGGCCAAAGUGGCUGCCAGAGCUGGAGCUGGAGAUGGAGCUGGAGC-----CUCAAAGGAGGAGGA------AA-AAAAGUGCAA------ ....(((((((((.((((.(((((((..(((.....)))..).)))))).)))))).........((....(-----(((....)))).)).------..-...)))))))------ ( -34.60) >consensus UUGAUUGCACUCAUGUUCAAGUUCUGUGGCCAAAGUGGCUGCCGGAGCUGGAG__________AGCUGGAGC_____CUCAAAGGAGGAGGAG___AAAA_AAAAGUGCAG______ ....(((((((........(((((((..(((.....)))..).))))))............................(((.......)))..............)))))))...... (-17.64 = -18.17 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:30 2006