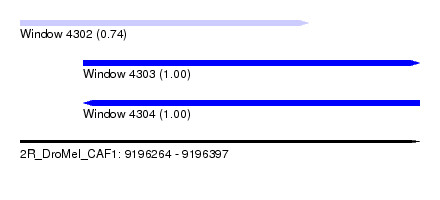
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,196,264 – 9,196,397 |
| Length | 133 |
| Max. P | 0.999703 |

| Location | 9,196,264 – 9,196,360 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9196264 96 + 20766785 U------UUAUAAAUCAUCAA---AUUGCC---AUUUUUU-AGCUGUUUGCCAUGUCAACAGGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGA---UGGAAUUUUC .------.......(((((..---((((((---.......-.((((((((.........)))))))).)))))).............(((....)))))---)))....... ( -26.30) >DroSec_CAF1 89676 96 + 1 U------GUAUAAAUCAUCAA---AUUGCC---AUUUUUU-AGCUGUUUGCCAUGUCAACAAGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGA---UGGAAUUCUC .------.......(((((..---((((((---.......-.((((((((.........)))))))).)))))).............(((....)))))---)))....... ( -26.70) >DroSim_CAF1 72818 96 + 1 U------GUAUAAAUCAUCAA---AUUGCC---AUUUUUU-AGCUGUUUGCCAUGUCAACAGGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGA---UGGAAUUUUC .------.......(((((..---((((((---.......-.((((((((.........)))))))).)))))).............(((....)))))---)))....... ( -26.30) >DroEre_CAF1 76720 97 + 1 U------GUAUAAAUCAUCAA---AUUUCC---AUUUUUUCAGCUGUUUGCCGUGUCAACGGGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGA---UGGAGCUUCC .------..............---..((((---((.......(((..(((((((....)).)))))..)))...............((((....)))))---)))))..... ( -26.80) >DroYak_CAF1 71958 96 + 1 U------GUAUAAAUCAUCAA---AUUUCC---AUUUUUU-AGCUGUUUGCCAUGUCAACAGGCAGUUGGCAAUAAAAUAUUAUUACCGCGGAAGCGGA---UGGAAUUUUC .------............((---(.((((---((.....-.(((..(((((.((....)))))))..)))...............((((....)))))---))))).))). ( -27.60) >DroPer_CAF1 62118 110 + 1 UUUAUACGUAUAAAUCAUCGGUGGAGGCCCGUACUUUUUUAGGCGGCUCGUAAUGGCAACAGGCAGUGGCCAAUAAAACAAUCGCCGUGCGGAAACAGCCAAUGA-AUUA-U .........((((.((((.(((......((((((.......(((((.......((....))(((....)))..........))))))))))).....))).))))-.)))-) ( -28.41) >consensus U______GUAUAAAUCAUCAA___AUUGCC___AUUUUUU_AGCUGUUUGCCAUGUCAACAGGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGA___UGGAAUUUUC .................(((......................(((..(((((.((....)))))))..)))...............((((....))))....)))....... (-15.75 = -16.08 + 0.34)

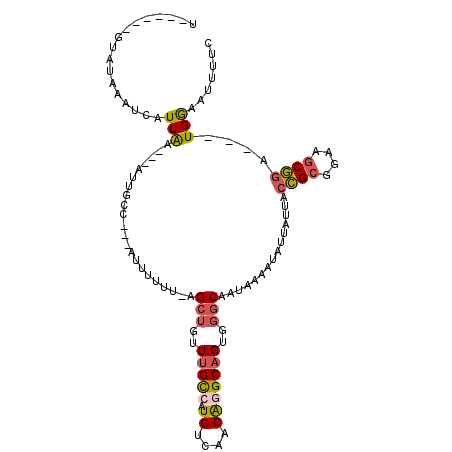
| Location | 9,196,285 – 9,196,397 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -37.41 |
| Consensus MFE | -30.78 |
| Energy contribution | -31.62 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9196285 112 + 20766785 AUUUUUU-AGCUGUUUGCCAUGUCAACAGGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGAUGGAAUUUUCUAUUCUUUGUGCUGUU---UUGUUUUGCCAGCAAACAGCC .......-.(((((((((((((((....)))).))((((((((((((.(((..((((....))))..(((((.....)))))..))).))))---))))..)))).))))))))). ( -39.40) >DroSec_CAF1 89697 112 + 1 AUUUUUU-AGCUGUUUGCCAUGUCAACAAGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGAUGGAAUUCUCUAUUCUUUGUGCUGUU---UGGUUUUUCCAGCAAACAGCC .......-.(((((((((........((((((((..((((.............((((....))))(((((....)))))...))))))))))---)).........))))))))). ( -37.63) >DroSim_CAF1 72839 112 + 1 AUUUUUU-AGCUGUUUGCCAUGUCAACAGGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGAUGGAAUUUUCUAUUCUUUGUGCUGUU---UGGUUUUUCCAGCAAACAGCC .......-.(((((((((........((((((((..((((.............((((....))))..(((((.....)))))))))))))))---)).........))))))))). ( -37.23) >DroEre_CAF1 76741 113 + 1 AUUUUUUCAGCUGUUUGCCGUGUCAACGGGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGAUGGAGCUUCCUAUUCUCUGUGUUUUU---GUGUUUUGCCAGCAAACAACC ...........((((((((((....)))(((((.(.((((..((((((.....((((....))))..((((.........))))))))))))---)).).))))).)))))))... ( -34.10) >DroYak_CAF1 71979 115 + 1 AUUUUUU-AGCUGUUUGCCAUGUCAACAGGCAGUUGGCAAUAAAAUAUUAUUACCGCGGAAGCGGAUGGAAUUUUCUAUUCUUUGUGUUGUUUCGCUGUUUUGCCAGCAAACAGCC .......-.((((((((((.((....))))).((((((((.............((((....))))..(((((.....)))))..(((......)))....))))))))))))))). ( -38.70) >consensus AUUUUUU_AGCUGUUUGCCAUGUCAACAGGCAGUGGGCAAUAAAAUAUUAUUACCGCGGAAGCGGAUGGAAUUUUCUAUUCUUUGUGCUGUU___UUGUUUUGCCAGCAAACAGCC .........(((((((((..((((....))))...(((((.............((((....))))..(((((.....)))))..................))))).))))))))). (-30.78 = -31.62 + 0.84)

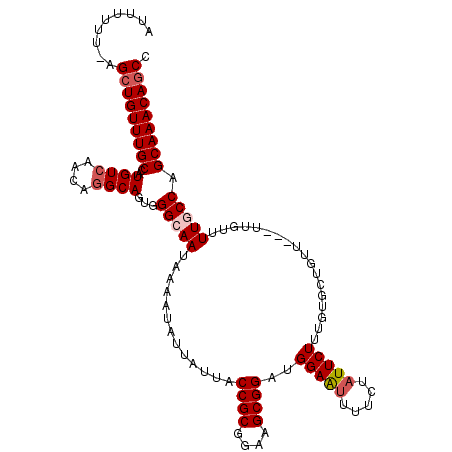
| Location | 9,196,285 – 9,196,397 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -28.34 |
| Energy contribution | -28.02 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.92 |
| SVM RNA-class probability | 0.999703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9196285 112 - 20766785 GGCUGUUUGCUGGCAAAACAA---AACAGCACAAAGAAUAGAAAAUUCCAUCCGCUUCCGCGGUAAUAAUAUUUUAUUGCCCACUGCCUGUUGACAUGGCAAACAGCU-AAAAAAU (((((((((((((((......---...(((.....((((.....)))).....)))...(.((((((((....)))))))))..))))((....)).)))))))))))-....... ( -31.90) >DroSec_CAF1 89697 112 - 1 GGCUGUUUGCUGGAAAAACCA---AACAGCACAAAGAAUAGAGAAUUCCAUCCGCUUCCGCGGUAAUAAUAUUUUAUUGCCCACUGCUUGUUGACAUGGCAAACAGCU-AAAAAAU (((((((((((((.....)).---..(((((...((.................((....))((((((((....))))))))..))...)))))....)))))))))))-....... ( -32.30) >DroSim_CAF1 72839 112 - 1 GGCUGUUUGCUGGAAAAACCA---AACAGCACAAAGAAUAGAAAAUUCCAUCCGCUUCCGCGGUAAUAAUAUUUUAUUGCCCACUGCCUGUUGACAUGGCAAACAGCU-AAAAAAU (((((((((((((.....)).---..(((((...((.................((....))((((((((....))))))))..))...)))))....)))))))))))-....... ( -32.30) >DroEre_CAF1 76741 113 - 1 GGUUGUUUGCUGGCAAAACAC---AAAAACACAGAGAAUAGGAAGCUCCAUCCGCUUCCGCGGUAAUAAUAUUUUAUUGCCCACUGCCCGUUGACACGGCAAACAGCUGAAAAAAU ((((((((((((....(((..---................((((((.......))))))(.((((((((....))))))))).......)))....))))))))))))........ ( -37.70) >DroYak_CAF1 71979 115 - 1 GGCUGUUUGCUGGCAAAACAGCGAAACAACACAAAGAAUAGAAAAUUCCAUCCGCUUCCGCGGUAAUAAUAUUUUAUUGCCAACUGCCUGUUGACAUGGCAAACAGCU-AAAAAAU (((((((((((.((......))....(((((...((.................((....))((((((((....))))))))..))...)))))....)))))))))))-....... ( -33.90) >consensus GGCUGUUUGCUGGCAAAACAA___AACAGCACAAAGAAUAGAAAAUUCCAUCCGCUUCCGCGGUAAUAAUAUUUUAUUGCCCACUGCCUGUUGACAUGGCAAACAGCU_AAAAAAU ((((((((((((....(((..................................((....))((((((((....))))))))........)))....))))))))))))........ (-28.34 = -28.02 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:28 2006