
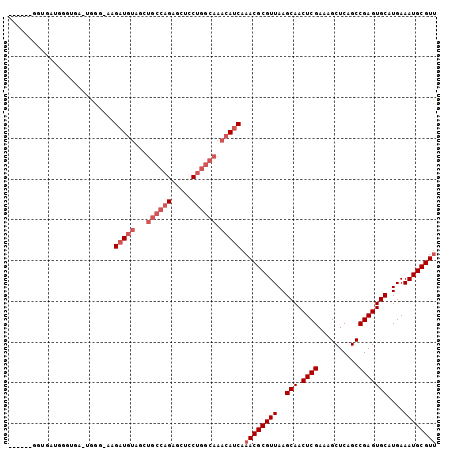
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,189,538 – 9,189,635 |
| Length | 97 |
| Max. P | 0.756570 |

| Location | 9,189,538 – 9,189,635 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -23.67 |
| Energy contribution | -25.17 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
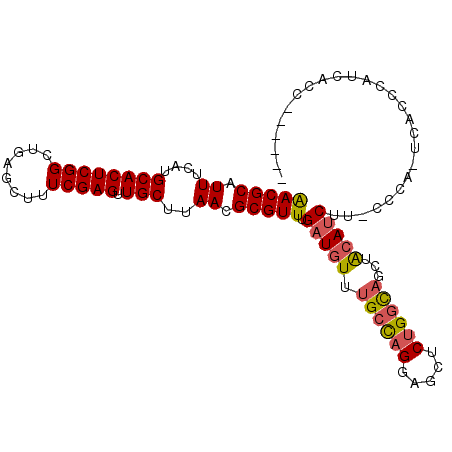
>2R_DroMel_CAF1 9189538 97 + 20766785 ------GGUUAUGGGUGA-UGGG-AAGAUGUAGCUGCCAGAGCUCCUGGCAAACAUCAAACGCGUUAAGCAACUCGAAAGCUCAGCCGAGUGCAUGAAAUGCGUU ------(((..(((((..-((((-..(((((...((((((.....)))))).)))))....((.....))..))))...))))))))....((((...))))... ( -29.90) >DroSec_CAF1 73858 103 + 1 GCCAUGGGUGAUGGGUGA-UGGG-AAGAUGUAGCUGCCAGAGCUCCUGGCAAACAUCAAACGCGUUAAGCAACUCGAAAGCUCAGCCGAGUGCAUGAAAUGCGUU .((((.(.(....).).)-))).-..(((((...((((((.....)))))).))))).((((((((..(((.((((..........)))))))....)))))))) ( -32.50) >DroSim_CAF1 66237 103 + 1 GCGAUGGGUGAUGGGUGA-UGGG-AAGAUGUAGCUGCCAGAGCUCCUGGCAAACAUCAAACGCGUUAAGCAACUCGAAAGCUCAGCCGAGUGCAUGAAAUGCGUU (((.(.(((..(((((..-((((-..(((((...((((((.....)))))).)))))....((.....))..))))...)))))))).).)))............ ( -31.90) >DroEre_CAF1 70557 85 + 1 -------------------GGGG-UAGAUGUAGCUGCCAGAGCUCCUGGCAAACAUCAAACGCGUUAAGCAACUCGAAAGCUCAGCCGAGUGCAUGAAAUGCGUU -------------------....-..(((((...((((((.....)))))).))))).((((((((..(((.((((..........)))))))....)))))))) ( -29.00) >DroYak_CAF1 65347 90 + 1 ----------AUGA-----GGGAUGAGAUGUAGCUGCCAGAGCUCCUGGCAAACAUCAAACGCGUUAAGCAACUCGAAAGCUCAGCCGAGUGCAUGAAAUGCGUU ----------....-----.......(((((...((((((.....)))))).))))).((((((((..(((.((((..........)))))))....)))))))) ( -29.00) >DroAna_CAF1 72610 102 + 1 GAGGAGGAAGAUAGUGGACUGGG-AGGCUCCAC--AUCUGAGCCACCAUCAAACAUCAAACGCGUUAAGCAACUCGAAAGCUCAGCCGAGUGCAUGAAAUGCGUC ((((.((.((((.(((((((...-.)).)))))--))))...)).)).)).........(((((((..(((.((((..........)))))))....))))))). ( -30.70) >consensus ______GGUGAUGGGUGA_UGGG_AAGAUGUAGCUGCCAGAGCUCCUGGCAAACAUCAAACGCGUUAAGCAACUCGAAAGCUCAGCCGAGUGCAUGAAAUGCGUU ..........................(((((...((((((.....)))))).))))).((((((((..(((.((((..........)))))))....)))))))) (-23.67 = -25.17 + 1.50)


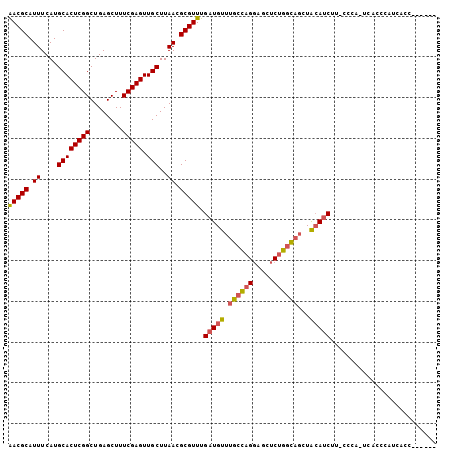
| Location | 9,189,538 – 9,189,635 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -23.37 |
| Energy contribution | -23.65 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9189538 97 - 20766785 AACGCAUUUCAUGCACUCGGCUGAGCUUUCGAGUUGCUUAACGCGUUUGAUGUUUGCCAGGAGCUCUGGCAGCUACAUCUU-CCCA-UCACCCAUAACC------ (((((.((....((((((((........))))).)))..)).))))).((((((((((((.....)))))))..)))))..-....-............------ ( -27.30) >DroSec_CAF1 73858 103 - 1 AACGCAUUUCAUGCACUCGGCUGAGCUUUCGAGUUGCUUAACGCGUUUGAUGUUUGCCAGGAGCUCUGGCAGCUACAUCUU-CCCA-UCACCCAUCACCCAUGGC (((((.((....((((((((........))))).)))..)).))))).((((((((((((.....)))))))..)))))..-....-....((((.....)))). ( -29.00) >DroSim_CAF1 66237 103 - 1 AACGCAUUUCAUGCACUCGGCUGAGCUUUCGAGUUGCUUAACGCGUUUGAUGUUUGCCAGGAGCUCUGGCAGCUACAUCUU-CCCA-UCACCCAUCACCCAUCGC (((((.((....((((((((........))))).)))..)).))))).((((((((((((.....)))))))..)))))..-....-.................. ( -27.30) >DroEre_CAF1 70557 85 - 1 AACGCAUUUCAUGCACUCGGCUGAGCUUUCGAGUUGCUUAACGCGUUUGAUGUUUGCCAGGAGCUCUGGCAGCUACAUCUA-CCCC------------------- (((((.((....((((((((........))))).)))..)).))))).((((((((((((.....)))))))..)))))..-....------------------- ( -27.30) >DroYak_CAF1 65347 90 - 1 AACGCAUUUCAUGCACUCGGCUGAGCUUUCGAGUUGCUUAACGCGUUUGAUGUUUGCCAGGAGCUCUGGCAGCUACAUCUCAUCCC-----UCAU---------- (((((.((....((((((((........))))).)))..)).))))).((((((((((((.....)))))))..))))).......-----....---------- ( -27.30) >DroAna_CAF1 72610 102 - 1 GACGCAUUUCAUGCACUCGGCUGAGCUUUCGAGUUGCUUAACGCGUUUGAUGUUUGAUGGUGGCUCAGAU--GUGGAGCCU-CCCAGUCCACUAUCUUCCUCCUC (((((.((....((((((((........))))).)))..)).))))).((((...(((((.(((((....--...))))).-))..)))...))))......... ( -27.10) >consensus AACGCAUUUCAUGCACUCGGCUGAGCUUUCGAGUUGCUUAACGCGUUUGAUGUUUGCCAGGAGCUCUGGCAGCUACAUCUU_CCCA_UCACCCAUCACC______ (((((.((....((((((((........))))).)))..)).))))).(((((.((((((.....))))))...))))).......................... (-23.37 = -23.65 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:23 2006