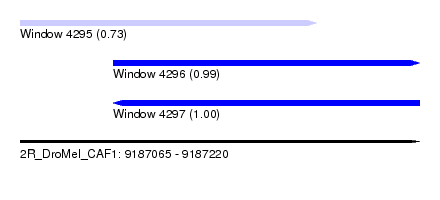
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,187,065 – 9,187,220 |
| Length | 155 |
| Max. P | 0.997542 |

| Location | 9,187,065 – 9,187,180 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.71 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.58 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
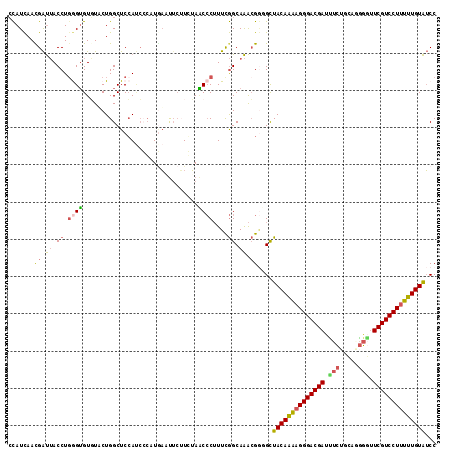
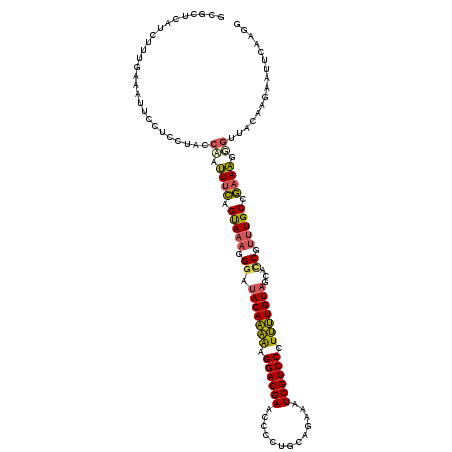
>2R_DroMel_CAF1 9187065 115 + 20766785 CCAUCAACGAUUACCUCAGUGUGUUCUGGCUCCAUCCCUUGAAUUCUUGUAACCCUUUCGGCAAACGGUGCUACAAAAGGGACGAUUUCUGCAGGGGUUCGUCCUUUUUGUAUCC .......(((......(((.(.((((.((.......))..)))).))))........)))(((.....)))(((((((((((((((..(....)..).))))))))))))))... ( -27.44) >DroSec_CAF1 71404 115 + 1 CCAUCAACGAUUACCUGGGUGUUUUCUGGCUCCAUCCCUUGAAUUCUUGUAACCCUUUCGGCAAACGGUGCUACAAAAGGGACGAUUUCAGCAUGGGUUCGUCCUUUUUGUAUCC .......(((......(((((..(((.((.......))..)))..).....))))..)))(((.....)))((((((((((((((.(((.....))).))))))))))))))... ( -29.30) >DroSim_CAF1 63750 115 + 1 CCAUCAAUGAUUACCUGGGUGUGUUCUGGCUCCAUCCCUUGAAUUCUUGUAACCCUUUCGGCAAACGGUGCUACAAAAGGGACGAUUUCUGCAUGGGUUCGUCCUUUUUGUAUCC ...........((((.((((..((((.((.......))..)))).......))))....(.....))))).((((((((((((((.(((.....))).))))))))))))))... ( -30.10) >DroEre_CAF1 68097 112 + 1 CCAUCAACGAUUACCUGGGUGUGUACUGGCUCCAUCGCAUCACU---UCUAGCCCUUUCGGCAAACGGGGCUACAAAAGGGACGAUCUCUGCAGGGGUUCGUCCUUUUUGUAUCC .............((((((((((...((....)).))))))...---....(((.....)))...))))..((((((((((((((((((....)))).))))))))))))))... ( -39.70) >DroYak_CAF1 62906 115 + 1 CCAUCAAUGAUUACCUGGGUGUGUACUGGCUCCAACGCAUAACUUCUUCUAGCCCUUUCGGCAAACGCGGUUACAAAAGGGACGAUCUCUGCAGGGUGUCGUCCUCUUUGUAUCC (((............)))((((((..((....))))))))...........(((.....)))......((.((((((..((((((((((....))).)))))))..)))))).)) ( -32.30) >DroAna_CAF1 69040 115 + 1 GUAUUAACGGCAACCUGAGGGUUUACUGGCUCGACGCGAUGUUCUCGUCGACCACAUUUAGCAGGAGAGGCUACACCAGGGACGAAUCAGGUAGAUAUUCGUCCUUGGUGUGGCC .............((((..((.....(((.((((((.(.....).)))))))))...))..))))...(((((((((((((((((((.........))))))))))))))))))) ( -50.90) >consensus CCAUCAACGAUUACCUGGGUGUGUACUGGCUCCAUCCCAUGAAUUCUUCUAACCCUUUCGGCAAACGGGGCUACAAAAGGGACGAUUUCUGCAGGGGUUCGUCCUUUUUGUAUCC .......((.((.((.((((...............................))))....)).)).))....((((((((((((((.(((.....))).))))))))))))))... (-22.77 = -22.58 + -0.19)

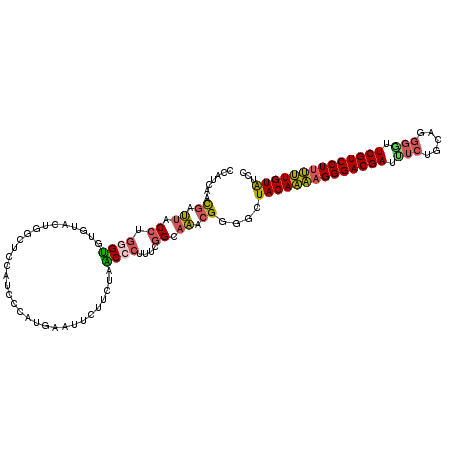
| Location | 9,187,101 – 9,187,220 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.48 |
| Mean single sequence MFE | -46.57 |
| Consensus MFE | -29.91 |
| Energy contribution | -30.58 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.38 |
| Mean z-score | -4.54 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
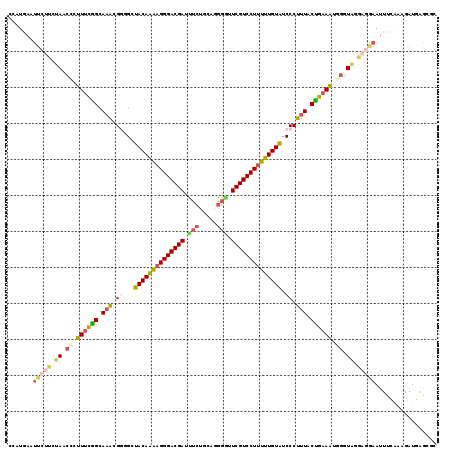
>2R_DroMel_CAF1 9187101 119 + 20766785 CCUUGAAUUCUUGUAACCCUUUCGGCAAACGGUGCUACAAAAGGGACGAUUUCUGCAGGGGUUCGUCCUUUUUGUAUCCCUUUACUGAAAUGGUUACGAGAAAUUUCAAAGAUGAGCGC ..((((((((((((((((.((((((.(((.((.(.(((((((((((((((..(....)..).)))))))))))))).)))))).)))))).)))))))))))..))))).......... ( -49.10) >DroSec_CAF1 71440 117 + 1 CCUUGAAUUCUUGUAACCCUUUCGGCAAACGGUGCUACAAAAGGGACGAUUUCAGCAUGGGUUCGUCCUUUUUGUAUCCCUUUACUGAAAUGGGUAGGAGGAAUUUUAAAGAUGGA--G .(((((((((((.(.((((((((((.(((.((.(.((((((((((((((.(((.....))).)))))))))))))).)))))).)))))).)))).).))))))))..))).....--. ( -44.30) >DroSim_CAF1 63786 117 + 1 CCUUGAAUUCUUGUAACCCUUUCGGCAAACGGUGCUACAAAAGGGACGAUUUCUGCAUGGGUUCGUCCUUUUUGUAUCCCUUUACUGAAAUGGGUAGGAGAAAUUUUGAAGAUGGA--A .(((.(((((((.((.(((((((((.(((.((.(.((((((((((((((.(((.....))).)))))))))))))).)))))).)))))).))))).)))))...)).))).....--. ( -42.00) >DroEre_CAF1 68133 116 + 1 GCAUCACU---UCUAGCCCUUUCGGCAAACGGGGCUACAAAAGGGACGAUCUCUGCAGGGGUUCGUCCUUUUUGUAUCCCUUUACUGAAAUUGGUGGAAGGAAUUGCACAGAUGACCGC (((...((---((((.(..((((((.(((.((((.((((((((((((((((((....)))).))))))))))))))))))))).))))))..).))))))....)))............ ( -48.20) >DroYak_CAF1 62942 119 + 1 GCAUAACUUCUUCUAGCCCUUUCGGCAAACGCGGUUACAAAAGGGACGAUCUCUGCAGGGUGUCGUCCUCUUUGUAUCCCUUUACCGAAAUUUGUAAAAGGAAUUCCUUAGAUGAGCGC ((.................((((((.(((.(.((.((((((..((((((((((....))).)))))))..)))))).)))))).)))))).(..(..((((....))))..)..))).. ( -37.10) >DroAna_CAF1 69076 119 + 1 CGAUGUUCUCGUCGACCACAUUUAGCAGGAGAGGCUACACCAGGGACGAAUCAGGUAGAUAUUCGUCCUUGGUGUGGCCCUAUGCUACAUUUGGUGGCACGAACUCCGACGACGAGUCU .......(((((((((((.((.(((((..((.(((((((((((((((((((.........))))))))))))))))))))).))))).)).))))((........))..)))))))... ( -58.70) >consensus CCAUGAAUUCUUCUAACCCUUUCGGCAAACGGGGCUACAAAAGGGACGAUUUCUGCAGGGGUUCGUCCUUUUUGUAUCCCUUUACUGAAAUGGGUAGGAGGAAUUUCAAAGAUGAGCGC .......(((((.((.((.((((((.(((.((...((((((((((((((.(((.....))).)))))))))))))).)).))).)))))).)).)).)))))................. (-29.91 = -30.58 + 0.68)


| Location | 9,187,101 – 9,187,220 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.48 |
| Mean single sequence MFE | -39.31 |
| Consensus MFE | -24.28 |
| Energy contribution | -23.95 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9187101 119 - 20766785 GCGCUCAUCUUUGAAAUUUCUCGUAACCAUUUCAGUAAAGGGAUACAAAAAGGACGAACCCCUGCAGAAAUCGUCCCUUUUGUAGCACCGUUUGCCGAAAGGGUUACAAGAAUUCAAGG ........(((((((..((((.((((((.((((.(((((.((.(((((((.((((((.............)))))).)))))))...)).))))).)))).)))))).))))))))))) ( -42.92) >DroSec_CAF1 71440 117 - 1 C--UCCAUCUUUAAAAUUCCUCCUACCCAUUUCAGUAAAGGGAUACAAAAAGGACGAACCCAUGCUGAAAUCGUCCCUUUUGUAGCACCGUUUGCCGAAAGGGUUACAAGAAUUCAAGG .--.....(((...(((((.....((((.((((.(((((.((.(((((((.((((((.............)))))).)))))))...)).))))).)))))))).....))))).))). ( -34.72) >DroSim_CAF1 63786 117 - 1 U--UCCAUCUUCAAAAUUUCUCCUACCCAUUUCAGUAAAGGGAUACAAAAAGGACGAACCCAUGCAGAAAUCGUCCCUUUUGUAGCACCGUUUGCCGAAAGGGUUACAAGAAUUCAAGG .--..............((((...((((.((((.(((((.((.(((((((.((((((.............)))))).)))))))...)).))))).))))))))....))))....... ( -33.92) >DroEre_CAF1 68133 116 - 1 GCGGUCAUCUGUGCAAUUCCUUCCACCAAUUUCAGUAAAGGGAUACAAAAAGGACGAACCCCUGCAGAGAUCGUCCCUUUUGUAGCCCCGUUUGCCGAAAGGGCUAGA---AGUGAUGC ((((....))))(((.(..((((..((..((((.((((((((.(((((((.((((((.((......).).)))))).)))))))..))).))))).)))).))...))---))..)))) ( -36.20) >DroYak_CAF1 62942 119 - 1 GCGCUCAUCUAAGGAAUUCCUUUUACAAAUUUCGGUAAAGGGAUACAAAGAGGACGACACCCUGCAGAGAUCGUCCCUUUUGUAACCGCGUUUGCCGAAAGGGCUAGAAGAAGUUAUGC ..((((....((((....)))).......(((((((((((((.(((((((.(((((((.(......).).)))))).))))))).)).).))))))))))))))............... ( -37.40) >DroAna_CAF1 69076 119 - 1 AGACUCGUCGUCGGAGUUCGUGCCACCAAAUGUAGCAUAGGGCCACACCAAGGACGAAUAUCUACCUGAUUCGUCCCUGGUGUAGCCUCUCCUGCUAAAUGUGGUCGACGAGAACAUCG ...(((((((..((........))((((....(((((.(((((.((((((.((((((((.........)))))))).)))))).))).))..)))))....)))))))))))....... ( -50.70) >consensus GCGCUCAUCUUUGAAAUUCCUCCUACCAAUUUCAGUAAAGGGAUACAAAAAGGACGAACCCCUGCAGAAAUCGUCCCUUUUGUAGCACCGUUUGCCGAAAGGGUUACAAGAAUUCAAGG ..........................((.((((.(((((.((.(((((((.((((((.............)))))).)))))))...)).))))).)))).))................ (-24.28 = -23.95 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:21 2006