
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,180,368 – 9,180,473 |
| Length | 105 |
| Max. P | 0.648664 |

| Location | 9,180,368 – 9,180,473 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.85 |
| Mean single sequence MFE | -42.91 |
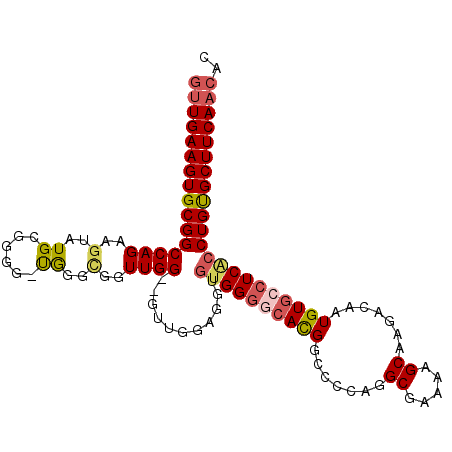
| Consensus MFE | -29.72 |
| Energy contribution | -30.39 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
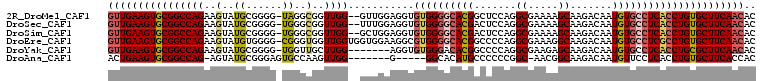
>2R_DroMel_CAF1 9180368 105 - 20766785 GUUGAAGUGCGGCCAGAAGUAUGCGGGG-UAGGCGGUUGG--GUUGGAGGUGUGGGGCACGGCUCCAGGCGAAAAGCAAGACAAUGUGCCUCACCUGUGCUUCAACAC (((((((..(((((((..((.(((...)-)).))..))))--.........((((((((((.......((.....)).......)))))))))))))..))))))).. ( -42.84) >DroSec_CAF1 64706 105 - 1 GUUGAAGUGCGGCCAGAAGUAUGCGGGG-UGGGCGGUUGG--UUUGGAGGUGUGGGGCACGACUCCAGGCGAAAAGCAAGACAAUGUGCCUCACCUGUGCUUCAACAC (((((((..((((((((....(((....-...))).....--)))))....((((((((((.......((.....)).......)))))))))))))..))))))).. ( -42.44) >DroSim_CAF1 57045 105 - 1 GUUGAAGUGCGGCCAGAAGUAUGCGGGG-UGGGCGGUUGG--GCUGGAGGUGUGGGGCACGACUCCAGGCGAAAAGCAAGACAAUGUGCCUCACCUGUGCUUCAACAC (((((((..(.((.........)).(((-((((((((((.--.((((((.((((...)))).))))))((.....))....)))).)))).))))))..))))))).. ( -44.10) >DroEre_CAF1 61433 107 - 1 GUUGAAGUGCGGCCAGAAGUAUGUGGGG-CGGGUGGUUGGUGGUGGAAGGCGUGGGGCACGGCCCCAGGCGAAAGGCAAGACAAUGUGCCUCGCCUGUGCUUCAACAC (((((((..(((..........((((((-((.((.(((..((.(.....((.((((((...)))))).))....).)).))).)).)))))))))))..))))))).. ( -48.40) >DroYak_CAF1 56075 100 - 1 GUUGAAGUGCGGCCAGAAGUAUGCGGGG-UGGUUGCUUGG-------AGGUGUGGGACACGGCCCCAGGCGAAGAGCAAGACAAUGUGCCUCACCUGCGCUUCAACAC ((((((((((((...((.(((((..(.(-(..((((((((-------.((((((...))).)))))))))))...))....)..))))).))..)))))))))))).. ( -41.50) >DroAna_CAF1 59095 94 - 1 ACUGAAGUGCGGCCAG-AGUAUGCGGGAGUGCCAAGUUGG-------G-----GGCACAUGCCCCCCGGC-AACGGCAAGACAAUGUUCCUCACCUGUGCUUCACCAC ..(((((..(((...(-((...(((....((((.....((-------(-----(((....)))))).(..-..)))))......)))..)))..)))..))))).... ( -38.20) >consensus GUUGAAGUGCGGCCAGAAGUAUGCGGGG_UGGGCGGUUGG__GUUGGAGGUGUGGGGCACGGCCCCAGGCGAAAAGCAAGACAAUGUGCCUCACCUGUGCUUCAACAC ((((((((((((((((..(..((......))..)..))))...........((((((((((.......((.....)).......)))))))))))))))))))))).. (-29.72 = -30.39 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:15 2006