| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,180,113 – 9,180,207 |
| Length | 94 |
| Max. P | 0.571886 |

| Location | 9,180,113 – 9,180,207 |
|---|---|
| Length | 94 |
| Sequences | 6 |
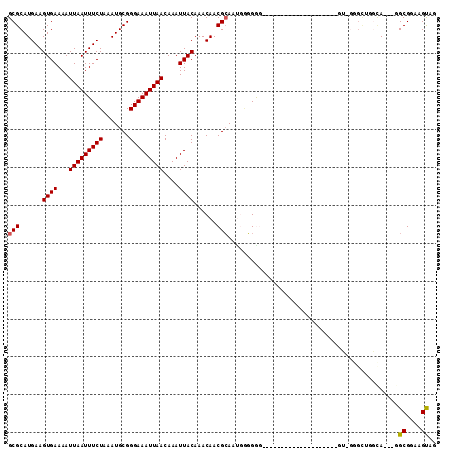
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -12.47 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.44 |
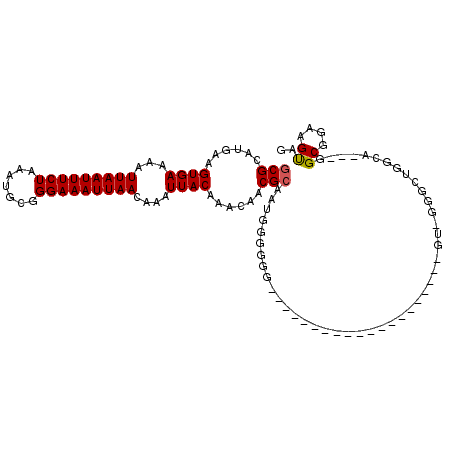
| Structure conservation index | 0.60 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
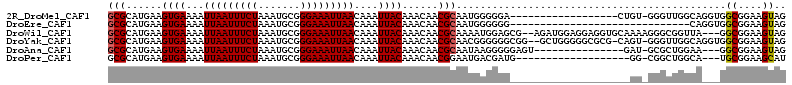
>2R_DroMel_CAF1 9180113 94 + 20766785 GCGCAUGAAGUGAAAAUUAAUUUCUAAAUGCGGGAAAUUAACAAAUUACAAACAACGCAAUGGGGGA------------------CUGU-GGGUUGGCAGGUGGCGGAAGUAG .(((.((..((((...(((((((((.......)))))))))....))))...((((.((.(((....------------------))))-).)))).)).)))((....)).. ( -23.30) >DroEre_CAF1 61191 83 + 1 GCGCAUGAAGUGAAAAUUAAUUUCUAAAUGCGGGAAAUUAACAAAUUACAAACAACGCAAUGGGGGG------------------------------CAGGUGGCGGAAGUAG .(((.....((((...(((((((((.......)))))))))....)))).......((........)------------------------------)..)))((....)).. ( -13.90) >DroWil_CAF1 80529 108 + 1 GCGCAUGAAGUGAAAAUUAAUUUCUAAAUGCGGGAAAUUAACAAAUUACAAACAACGCAAAAUGGAGCG--AGAUGGAGGAGGUGCAAAAGGGCGGUUA---GGCGGAAGUAG ((((.....((((...(((((((((.......)))))))))....))))......(((........)))--...........)))).............---.((....)).. ( -19.60) >DroYak_CAF1 55639 109 + 1 GCGCAUGAAGUGAAAAUUAAUUUCUAAAUGCGGGAAAUUAACAAAUUACAAACAACGCAACGGGGGGCGG--GCUGGGGGCGCG-CAGU-GGGUUGGCAGGUGGCGGAAGUAG .(((.((..((((...(((((((((.......)))))))))....))))...((((.((.(.....(((.--((.....)).))-).))-).)))).)).)))((....)).. ( -27.80) >DroAna_CAF1 58885 94 + 1 GCGCAUGAAGUGAAAAUUAAUUUCUAAAUGCGGGAAAUUAACAAAUUACAAACAACGCAAUAAGGGGGAGU---------------GAU-GCGCUGGAA---GGCGGAAGUAG ((((((...((((...(((((((((.......)))))))))....))))..((..(.(......).)..))---------------.))-)))).....---.((....)).. ( -20.50) >DroPer_CAF1 48282 90 + 1 GCGCAUGAAGUGAAAAUUAAUUUCUAAAUGCGGGAAAUUAACAAAUUACAAACAACGGAAUGACGAUG-------------------GG-CGGCUGGCA---UGCGGAAGCAU ((.(((...((((...(((((((((.......)))))))))....))))......((......)))))-------------------.)-).......(---(((....)))) ( -20.50) >consensus GCGCAUGAAGUGAAAAUUAAUUUCUAAAUGCGGGAAAUUAACAAAUUACAAACAACGCAAUGGGGGG____________________GU_GGGCUGGCA___GGCGGAAGUAG (((......((((...(((((((((.......)))))))))....))))......))).............................................((....)).. (-12.47 = -12.50 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:14 2006