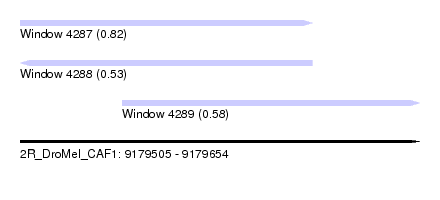
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,179,505 – 9,179,654 |
| Length | 149 |
| Max. P | 0.823656 |

| Location | 9,179,505 – 9,179,614 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.20 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.07 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9179505 109 + 20766785 AGACUACUUGAGUCAAUUUAAAAAGAGA--UUUCUUCGGAACAUAGAAGAGAAC-CCUGUGCCAUCUUUAUUUC--------GGGGUGUUCUAGCCCUUGGGGCGAUUCAAGUGCCUUGG ....((((((((((.........((((.--(((((((........)))))))((-((((..............)--------))))).)))).((((...))))))))))))))...... ( -34.64) >DroSec_CAF1 63811 116 + 1 AGACUACUUGAGUCAAUUUAAAAAGAGA--UUUCUUCGGUACUUCUAAUAGUAC-CCUGCACCAUCUUUAUAUAUUAUUCUUUAUAUGUUGUAGCCCUUG-GGCGAUUCAAGUGCCUUGG ....((((((((((.........(((((--(......((((((......)))))-).......))))))((((((........))))))....(((....-)))))))))))))...... ( -29.52) >DroSim_CAF1 56150 110 + 1 AGACUACUUGAGUCAAUUUAAAAAGAUAUAUGUCUUUGGUACUUCUAAUAGUAC-UUUGCACCAUCUUUAUAUC--------AGGGUGUUGUAGCCCUUG-GGCGAUUCAAGUGCCUUGG ....((((((((((.......((((((....))))))((((((......)))))-).((((.((((((......--------)))))).))))(((....-)))))))))))))...... ( -33.30) >DroEre_CAF1 60595 109 + 1 AGACUACUUGAGUCAAUUUAAAAAGACA--UUUCUUUGUUACACCCAAUAUAGC-CCUGCACUAUCUUUUUAGC--------AGUGUGUUGUAGCCCUUGGGCCCAUUGAAGUGCCUUGG .((((.....))))........(((.((--((((..((..........((((((-(((((............))--------)).).))))))(((....))).))..)))))).))).. ( -26.20) >DroYak_CAF1 55006 105 + 1 AGACUACUUGAGUCAGUUCGAAAAGAGC--UUUCUUAGGUACAUCCAAUAGAAUCUAUGCUCUAUCUUCAUA-------------GUGUGGUAGCUCUUGGGGCCAUUCAAGUGCCUUGG ....((((((((..(((((.....))))--)..))))))))...((((.((...(((((((..........)-------------))))))...)).))))((((......).))).... ( -27.50) >consensus AGACUACUUGAGUCAAUUUAAAAAGAGA__UUUCUUCGGUACAUCCAAUAGAAC_CCUGCACCAUCUUUAUAUC________AGGGUGUUGUAGCCCUUGGGGCGAUUCAAGUGCCUUGG .((((.....))))........(((((....))))).(((((...............((((.(((....................))).))))(((.....))).......))))).... (-13.31 = -13.07 + -0.24)

| Location | 9,179,505 – 9,179,614 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.20 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -14.48 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9179505 109 - 20766785 CCAAGGCACUUGAAUCGCCCCAAGGGCUAGAACACCCC--------GAAAUAAAGAUGGCACAGG-GUUCUCUUCUAUGUUCCGAAGAAA--UCUCUUUUUAAAUUGACUCAAGUAGUCU ...((((((((((.((((((...))))..((((.((((--------(.........)))....))-))))(((((........)))))..--..............)).)))))).)))) ( -25.30) >DroSec_CAF1 63811 116 - 1 CCAAGGCACUUGAAUCGCC-CAAGGGCUACAACAUAUAAAGAAUAAUAUAUAAAGAUGGUGCAGG-GUACUAUUAGAAGUACCGAAGAAA--UCUCUUUUUAAAUUGACUCAAGUAGUCU ...((((((((((.(((((-....))).................(((.((.(((((.(((....(-(((((......))))))......)--))))))).)).))))).)))))).)))) ( -28.30) >DroSim_CAF1 56150 110 - 1 CCAAGGCACUUGAAUCGCC-CAAGGGCUACAACACCCU--------GAUAUAAAGAUGGUGCAAA-GUACUAUUAGAAGUACCAAAGACAUAUAUCUUUUUAAAUUGACUCAAGUAGUCU ...((((((((((.(((((-....)))...........--------((((((..((((((((...-))))))))....((.......)).))))))..........)).)))))).)))) ( -26.40) >DroEre_CAF1 60595 109 - 1 CCAAGGCACUUCAAUGGGCCCAAGGGCUACAACACACU--------GCUAAAAAGAUAGUGCAGG-GCUAUAUUGGGUGUAACAAAGAAA--UGUCUUUUUAAAUUGACUCAAGUAGUCU ..((((((.(((..((.((((((.((((......((((--------(((....)).)))))...)-)))...))))))....))..))).--))))))........((((.....)))). ( -27.50) >DroYak_CAF1 55006 105 - 1 CCAAGGCACUUGAAUGGCCCCAAGAGCUACCACAC-------------UAUGAAGAUAGAGCAUAGAUUCUAUUGGAUGUACCUAAGAAA--GCUCUUUUCGAACUGACUCAAGUAGUCU ...((((((((((..(..(..(((((((......(-------------((((.........))))).((((...((.....))..)))))--))))))...)..)....)))))).)))) ( -23.20) >consensus CCAAGGCACUUGAAUCGCCCCAAGGGCUACAACACACU________GAUAUAAAGAUGGUGCAGG_GUUCUAUUAGAUGUACCAAAGAAA__UCUCUUUUUAAAUUGACUCAAGUAGUCU ...((((((((((...(((.....)))...........................((((((((....)))))))).........(((((......)))))..........)))))).)))) (-14.48 = -15.40 + 0.92)

| Location | 9,179,543 – 9,179,654 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -35.19 |
| Consensus MFE | -15.20 |
| Energy contribution | -16.92 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9179543 111 + 20766785 ACAUAGAAGAGAAC-CCUGUGCCAUCUUUAUUUC--------GGGGUGUUCUAGCCCUUGGGGCGAUUCAAGUGCCUUGGCCCAAUUCUUCGCGAAUGUGGGCUGAGCGAUUCUGUGAUU .((((((((((.((-((((..............)--------))))).)))).((((...))))........(((.(..(((((((((.....)))).)))))..))))..))))))... ( -39.64) >DroSec_CAF1 63849 118 + 1 ACUUCUAAUAGUAC-CCUGCACCAUCUUUAUAUAUUAUUCUUUAUAUGUUGUAGCCCUUG-GGCGAUUCAAGUGCCUUGGCCCAAUUCCUCGCGAGAGUGGGCUGAGCGAUUCUGUGAUU .......((((...-...((((......((((((........))))))(((..(((....-)))....)))))))((..(((((....(((....))))))))..)).....)))).... ( -31.80) >DroSim_CAF1 56190 110 + 1 ACUUCUAAUAGUAC-UUUGCACCAUCUUUAUAUC--------AGGGUGUUGUAGCCCUUG-GGCGAUUCAAGUGCCUUGGCCCAAUUCCUCGCGAAAGUGGGCUGAGCGAUUCUGUGAUU ..........((((-((((((.((((((......--------)))))).))))(((....-))).....))))))((..(((((.(((.....)))..)))))..))............. ( -32.20) >DroEre_CAF1 60633 109 + 1 ACACCCAAUAUAGC-CCUGCACUAUCUUUUUAGC--------AGUGUGUUGUAGCCCUUGGGCCCAUUGAAGUGCCUUGGCCCAUUUCUUCGCGAAAGUGGGCUG--CGACGCUGUGAUU ........((((((-...(((((..((....)).--------)))))((((((((((.(((((((((....)))....)))))).......((....))))))))--))))))))))... ( -41.60) >DroYak_CAF1 55044 105 + 1 ACAUCCAAUAGAAUCUAUGCUCUAUCUUCAUA-------------GUGUGGUAGCUCUUGGGGCCAUUCAAGUGCCUUGGCCCAAUUCUUCGCGUAACUGGGCUG--CGAUUCUGUGAUU .......((((((((...((.((((....)))-------------).)).(((((((...((((((...........))))))................))))))--))))))))).... ( -30.71) >consensus ACAUCCAAUAGAAC_CCUGCACCAUCUUUAUAUC________AGGGUGUUGUAGCCCUUGGGGCGAUUCAAGUGCCUUGGCCCAAUUCUUCGCGAAAGUGGGCUGAGCGAUUCUGUGAUU ...................(((.(((...........................((.((((((....)))))).))(((((((((..((.....))...))))))))).)))...)))... (-15.20 = -16.92 + 1.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:13 2006