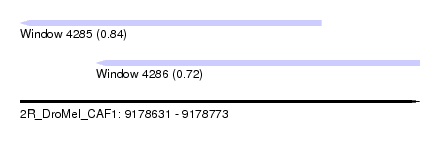
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,178,631 – 9,178,773 |
| Length | 142 |
| Max. P | 0.843277 |

| Location | 9,178,631 – 9,178,738 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -20.42 |
| Energy contribution | -21.53 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9178631 107 - 20766785 UUGUAAAGUGCAUGCUUUCUGGCAUACAGAAGGCGUGUAUUGUUCACGCCGAGAAGAUUGUCUACCAGAAGAAGUUCGCUGAACAAAUCGCGAU------U---GAGGAGGCUAAA---- .((...(((((((((((((((.....)))))))))))))))...)).(((.....((((((((......))).((((...))))......))))------)---.....)))....---- ( -32.50) >DroSec_CAF1 62935 107 - 1 UUGUAAAGUGCAUGCUUUCUGGCAUACAGAAGGCGUGUGUUGUUCAUGCCGAGAAGAUGGUCUACCAGAAGAAGUUCGCUGAACAAAUUGCGAU------U---GAGGAGGCUAUA---- ((((((...((((((((((((.....)))))))))))).(((((((.((.(((....(((....))).......)))))))))))).)))))).------.---............---- ( -34.20) >DroSim_CAF1 55274 107 - 1 UUGUAAAGUGCAUGCUUUCUGGCAUACAGAAGGCGUGUGUUGUUCAUGCCGAGAAGAUGGUCUACCAGAAAAAGUUCGCUGAACAAAUUGCGAU------U---GAGGAGGCUAUA---- ((((((...((((((((((((.....)))))))))))).(((((((.((.(((....(((....))).......)))))))))))).)))))).------.---............---- ( -34.20) >DroEre_CAF1 59730 107 - 1 UUGUAAAGUGCAUGCUUUCUGGCAUAAAGAAGGCGUGUGUUGUUCACGCCGAGAAGAUUGUCUACCAGCAGAAGUUCGCUGAACAAAUUGCGGC------U---GAGGAGGCGGAG---- ((((..((..((((((((((.......))))))))))..)).((((.((((...((.((((....((((........)))).)))).)).))))------)---)))...))))..---- ( -32.60) >DroYak_CAF1 54147 107 - 1 UUGUAAAGUGCAUGCUUUCUGGCAUAAAGAAGGCGUGUACUGUGCAUGCCGAGAAGCUUGUCUACCAGCAGAAGUUUGCUGAACAAAUUGCGGC------U---GAGGAGGCAGAG---- .((((.((((((((((((((.......)))))))))))))).))))((((.(((......)))..((((...((((((.....))))))...))------)---)....))))...---- ( -40.50) >DroAna_CAF1 57496 120 - 1 UGGUCCGCUGCAUGCUGAAGGGCAUUAAGAAGGGCUGCCUGCUCCACGCCCAGAAGAAGAUCUGCGCCCAGAAGGACGCCAAGCAGAAUGCGGCAGCCAUUGCCGCCGAGGCGGAGAAGG ((((..(((((((.(((...(((........((((.....)).))..((.((((......)))).))((....))..)))...))).))))))).))))...((((....))))...... ( -44.30) >consensus UUGUAAAGUGCAUGCUUUCUGGCAUAAAGAAGGCGUGUGUUGUUCACGCCGAGAAGAUGGUCUACCAGAAGAAGUUCGCUGAACAAAUUGCGAC______U___GAGGAGGCUAAA____ .((...((((((((((((((.......))))))))))))))...)).(((.(((......)))............((((..........))))................)))........ (-20.42 = -21.53 + 1.11)


| Location | 9,178,658 – 9,178,773 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -23.33 |
| Energy contribution | -24.72 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9178658 115 - 20766785 AAAGCCUCA-----GGGUGAUCCCAAGUCUGUAGAUUCUCUUGUAAAGUGCAUGCUUUCUGGCAUACAGAAGGCGUGUAUUGUUCACGCCGAGAAGAUUGUCUACCAGAAGAAGUUCGCU ..(((....-----(((....)))...(((((((((..((((....(((((((((((((((.....))))))))))))))).(((.....)))))))..))))).))))........))) ( -39.40) >DroSec_CAF1 62962 115 - 1 AAAGCCGAA-----GGGUGAUCCCAAGGCCGUAGAUUCUCUUGUAAAGUGCAUGCUUUCUGGCAUACAGAAGGCGUGUGUUGUUCAUGCCGAGAAGAUGGUCUACCAGAAGAAGUUCGCU ..(((.(((-----(((....))).(((((((...(((((.((...((..(((((((((((.....)))))))))))..))...))....))))).)))))))...........)))))) ( -43.40) >DroSim_CAF1 55301 115 - 1 AAAGCCGAA-----GGGUGAUCCCAAGGCCGUAGAUUCUCUUGUAAAGUGCAUGCUUUCUGGCAUACAGAAGGCGUGUGUUGUUCAUGCCGAGAAGAUGGUCUACCAGAAAAAGUUCGCU ..(((.(((-----(((....))).(((((((...(((((.((...((..(((((((((((.....)))))))))))..))...))....))))).)))))))...........)))))) ( -43.40) >DroEre_CAF1 59757 115 - 1 GAAGCAGCA-----GGGGGAUCCCAAGUCCGUAGAUUCUCUUGUAAAGUGCAUGCUUUCUGGCAUAAAGAAGGCGUGUGUUGUUCACGCCGAGAAGAUUGUCUACCAGCAGAAGUUCGCU ..((((((.-----(((....)))..((..((((((..((((....((..((((((((((.......))))))))))..)).(((.....)))))))..))))))..))....))).))) ( -35.80) >DroYak_CAF1 54174 115 - 1 AAAGCCACA-----GGGUGAUCCCAAGUCCGUAGAUUCACUUGUAAAGUGCAUGCUUUCUGGCAUAAAGAAGGCGUGUACUGUGCAUGCCGAGAAGCUUGUCUACCAGCAGAAGUUUGCU ..(((.((.-----(((....)))..((..((((((.....((((.((((((((((((((.......)))))))))))))).)))).((......))..))))))..))....))..))) ( -37.50) >DroAna_CAF1 57536 120 - 1 GAAAAGGAAUUCUCGCACGAUCCCCCCGCCGUGGACAAGCUGGUCCGCUGCAUGCUGAAGGGCAUUAAGAAGGGCUGCCUGCUCCACGCCCAGAAGAAGAUCUGCGCCCAGAAGGACGCC .....(...((((((((.((((.....((.((((((......)))))).))........((((....((.(((....))).))....)))).......))))))))...))))...)... ( -35.50) >consensus AAAGCCGAA_____GGGUGAUCCCAAGGCCGUAGAUUCUCUUGUAAAGUGCAUGCUUUCUGGCAUAAAGAAGGCGUGUGUUGUUCACGCCGAGAAGAUGGUCUACCAGAAGAAGUUCGCU ..(((.........(((....)))......((((((..((((.(.......(((((....)))))......((((((.......)))))).).))))..))))))............))) (-23.33 = -24.72 + 1.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:11 2006