| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,173,906 – 9,174,005 |
| Length | 99 |
| Max. P | 0.603434 |

| Location | 9,173,906 – 9,174,005 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -18.60 |
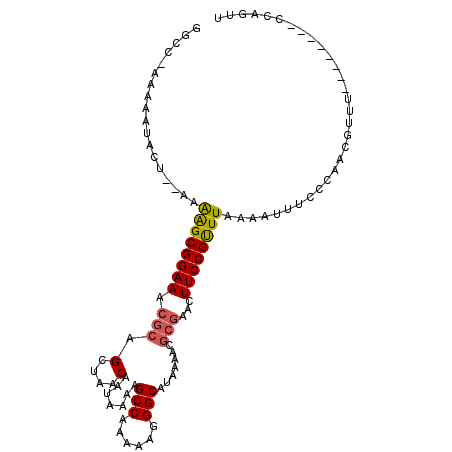
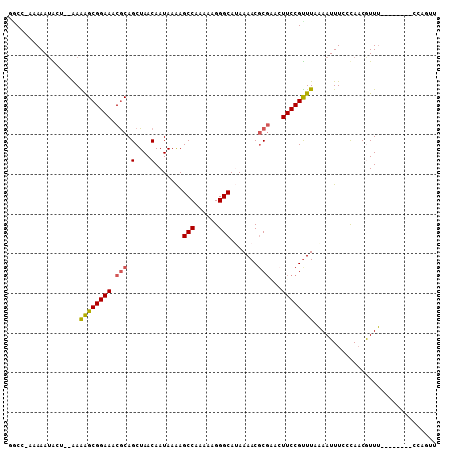
| Consensus MFE | -13.49 |
| Energy contribution | -13.47 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9173906 99 + 20766785 GGCC-AAAAAUACCAGAAAAGCGGAAACACAGCUAACAAUAAGAGCCGAAAAGGGCAGAAAACGCGAACUUCCGUUUAAAAUUUCCUAACAUUU--------CAAGCU ....-.............((((((((.....((...(.....).(((......))).......))....)))))))).................--------...... ( -13.60) >DroVir_CAF1 71782 105 + 1 UACC-AAAAAUGCU--GAGAGCGGAAACGCAGCUAACAAUAAAAGCCAAAGAGGGCAUAAAACGCGGAGUUCCGUUUUAAAUUUCCCAACGUUUCAUUCGUUCCGUUU ....-.........--..((((((((.((.....(((.......(((......))).......((((....))))...............))).....)))))))))) ( -21.20) >DroPse_CAF1 42616 99 + 1 GCCC-CCAAAAACACGAAAGGCGGAAACGCAGCUAACAAUAAAAGCCAAAAAGGGCAUAAAACUCCAACUUCCGUUUAGAAUUUCGCAACGUUC--------CGUUUU ((((-.........(....)(((....))).(((.........)))......))))................((((..(.....)..))))...--------...... ( -17.90) >DroGri_CAF1 61644 98 + 1 UACCGAAAAAUGUU--AAGAGCGGAAACGCAGCUAACAAUAAAAGCCAAAAAGGGCAUAAAACGCGCACUUCCGUUUUAAAUUUCCCAUCAUUU--------ACAGGU .(((...(((((..--..(((((....)))..............(((......))).(((((((........)))))))....))....)))))--------...))) ( -21.30) >DroMoj_CAF1 76496 97 + 1 UGCC-AAAAAUGCU--AAGAGCGGAAACGCAGCUAACAAUAAAAGCCAAAAAGGGCAUAAAACGCGCACUUCCGCUUUAAAUUUCCCAACGUUU--------UCACUU ....-.........--.(((((((((.(((.(....).......(((......))).......)))...)))))))))................--------...... ( -18.70) >DroAna_CAF1 52268 99 + 1 GGCC-AAAAAUACCGAAAAAGCGGAAACGCAGCUAACAAUAAAAGCCGAAAAGGGCAGAAAACGCGAGCUUCCGUUUAAAAUUUCCUAACGUUU--------CAAGCU ....-...............(((....)))(((((((.......(((......)))...(((((.(.....)))))).............))).--------..)))) ( -18.90) >consensus GGCC_AAAAAUACU__AAAAGCGGAAACGCAGCUAACAAUAAAAGCCAAAAAGGGCAUAAAACGCGAACUUCCGUUUAAAAUUUCCCAACGUUU________CCAGUU ..................((((((((.(((.(....).......(((......))).......)))...))))))))............................... (-13.49 = -13.47 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:09 2006