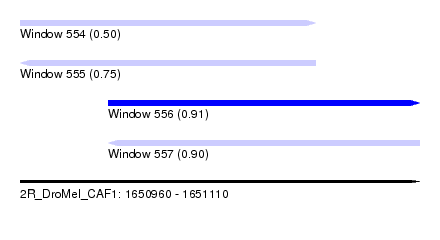
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,650,960 – 1,651,110 |
| Length | 150 |
| Max. P | 0.906000 |

| Location | 1,650,960 – 1,651,071 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.41 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -17.13 |
| Energy contribution | -18.13 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1650960 111 + 20766785 ----GUUUUUUGAAACAGUGAUUUGAAAUGCUUUGUUCUCAUUUCUCUCAUACAUACCCACAUGUAAAUAAACGUAUUGGUUUAAUGACACAUGCAGCUUAUUUGUAUGUGUUAU ----((((....)))).((((...((((((.........))))))..))))((((......))))...(((((......)))))(((((((((((((.....))))))))))))) ( -22.70) >DroSec_CAF1 36847 115 + 1 UUUUUUUUUUUGAUACAGUGAUUUGAAAUGCUUUGUUCUCAUUUCUCUCAUACAUACCCACAUGUAAAUAAACGUAUUGGUUUAAUGACACAUGCAGCUUGUUUGUAUGUGUUCU .........(..((((.((((...((((((.........))))))..))))((((......))))........))))..)......(((((((((((.....))))))))))).. ( -23.10) >DroEre_CAF1 40531 107 + 1 UUUUUUUUUUUGAUACAGUGAUUUGAAAUGCUUUGUUCUCAUUUCUCUCA--------CACAUGUAAAUAAAAGUAUUGGUUUAAUGACACAUGCAGUUUAUUUGUAUGUGUACG .........(..((((.((((...((((((.........))))))..)))--------)..((....))....))))..).......((((((((((.....))))))))))... ( -22.30) >consensus UUUUUUUUUUUGAUACAGUGAUUUGAAAUGCUUUGUUCUCAUUUCUCUCAUACAUACCCACAUGUAAAUAAACGUAUUGGUUUAAUGACACAUGCAGCUUAUUUGUAUGUGUUCU .........(..((((.((((...((((((.........))))))..))))..........((....))....))))..)......(((((((((((.....))))))))))).. (-17.13 = -18.13 + 1.00)

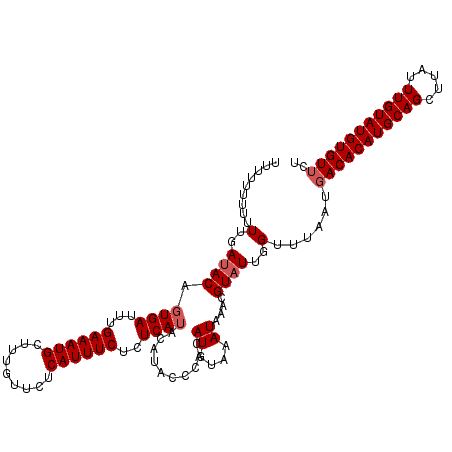
| Location | 1,650,960 – 1,651,071 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 88.41 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -15.40 |
| Energy contribution | -16.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1650960 111 - 20766785 AUAACACAUACAAAUAAGCUGCAUGUGUCAUUAAACCAAUACGUUUAUUUACAUGUGGGUAUGUAUGAGAGAAAUGAGAACAAAGCAUUUCAAAUCACUGUUUCAAAAAAC---- ..((((((((((....(.(..((((((....(((((......)))))..))))))..).).))))))...((((((.........)))))).......)))).........---- ( -22.10) >DroSec_CAF1 36847 115 - 1 AGAACACAUACAAACAAGCUGCAUGUGUCAUUAAACCAAUACGUUUAUUUACAUGUGGGUAUGUAUGAGAGAAAUGAGAACAAAGCAUUUCAAAUCACUGUAUCAAAAAAAAAAA .(((((((((((.((...(..((((((....(((((......)))))..))))))..))).))))))...((((((.........)))))).......))).))........... ( -23.80) >DroEre_CAF1 40531 107 - 1 CGUACACAUACAAAUAAACUGCAUGUGUCAUUAAACCAAUACUUUUAUUUACAUGUG--------UGAGAGAAAUGAGAACAAAGCAUUUCAAAUCACUGUAUCAAAAAAAAAAA .......(((((........(((((((....((((........))))..)))))))(--------(((..((((((.........))))))...)))))))))............ ( -16.80) >consensus AGAACACAUACAAAUAAGCUGCAUGUGUCAUUAAACCAAUACGUUUAUUUACAUGUGGGUAUGUAUGAGAGAAAUGAGAACAAAGCAUUUCAAAUCACUGUAUCAAAAAAAAAAA .......(((((......(((((((((....(((((......)))))..))))))))).......(((..((((((.........))))))...))).)))))............ (-15.40 = -16.40 + 1.00)

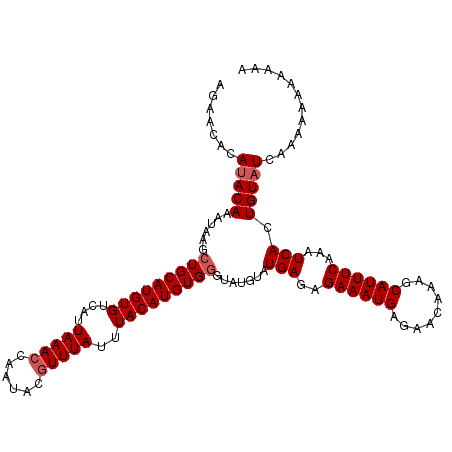
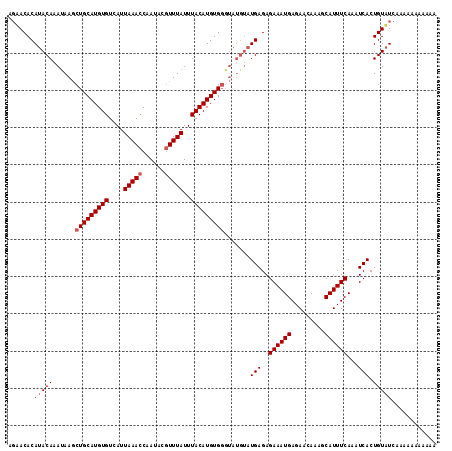
| Location | 1,650,993 – 1,651,110 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.95 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -22.23 |
| Energy contribution | -24.60 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.906000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1650993 117 + 20766785 CUCAUUUCUCUCAUACAUACCCACAUGUAAAUAAACGUAUUGGUUUAAUGACACAUGCAGCUUAUUUGUAUGUGUUAU-UUUUUUGGCAAUUUCACAUCCAGGUGUGUAUGUAUUAUC ...........((((((((((...((((...(((((......)))))(((((((((((((.....)))))))))))))-...............))))...))))))))))....... ( -31.90) >DroSec_CAF1 36884 118 + 1 CUCAUUUCUCUCAUACAUACCCACAUGUAAAUAAACGUAUUGGUUUAAUGACACAUGCAGCUUGUUUGUAUGUGUUCUGUUUUUUGGCCAUUUCACAUCCAGGUGUGUAUGUAUUAUC ...........((((((((((...((((......))))..(((((.((((((((((((((.....)))))))))))..)))....)))))...........))))))))))....... ( -30.10) >DroSim_CAF1 33762 118 + 1 CUCAUUUCUCUCAUACAUGCCCACAUGUAAAUAAACGUAUUGGUUUAAUGACACAUGCAGCUUGUUUGUAUGUGUUCUGUUUUUUGGCAAUUUCACAUCCAGGUGUGUAUGUAUUAUC ...........((((((((((...((((...(((((......)))))..(((((((((((.....))))))))))).((((....)))).....))))...))))))))))....... ( -29.00) >DroEre_CAF1 40568 110 + 1 CUCAUUUCUCUCA--------CACAUGUAAAUAAAAGUAUUGGUUUAAUGACACAUGCAGUUUAUUUGUAUGUGUACGGUUUUUUGCCAGUUUCACAUCCAGGUGUGUAUGUAUUAUC .............--------.(((((...........((((((..(((.((((((((((.....))))))))))...)))....))))))..(((((....))))))))))...... ( -24.60) >consensus CUCAUUUCUCUCAUACAUACCCACAUGUAAAUAAACGUAUUGGUUUAAUGACACAUGCAGCUUAUUUGUAUGUGUUCUGUUUUUUGGCAAUUUCACAUCCAGGUGUGUAUGUAUUAUC ...........((((((((((...((((...(((((......)))))..(((((((((((.....)))))))))))..................))))...))))))))))....... (-22.23 = -24.60 + 2.37)

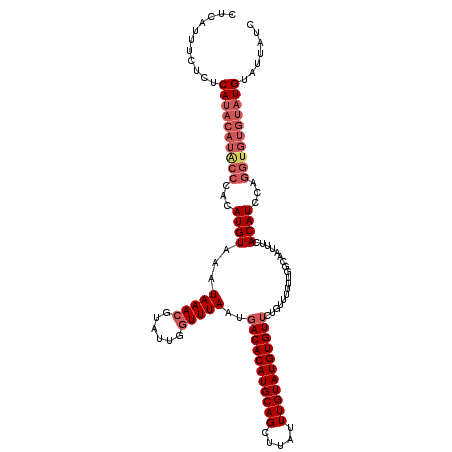
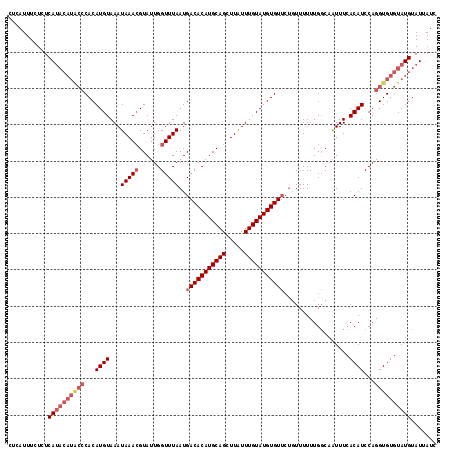
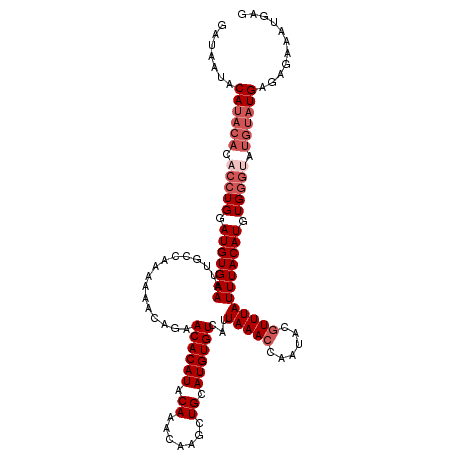
| Location | 1,650,993 – 1,651,110 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.95 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -20.12 |
| Energy contribution | -22.38 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1650993 117 - 20766785 GAUAAUACAUACACACCUGGAUGUGAAAUUGCCAAAAAA-AUAACACAUACAAAUAAGCUGCAUGUGUCAUUAAACCAAUACGUUUAUUUACAUGUGGGUAUGUAUGAGAGAAAUGAG .......((((((.(((((.(((((((............-...((((((.((.......)).))))))...(((((......)))))))))))).))))).))))))........... ( -27.30) >DroSec_CAF1 36884 118 - 1 GAUAAUACAUACACACCUGGAUGUGAAAUGGCCAAAAAACAGAACACAUACAAACAAGCUGCAUGUGUCAUUAAACCAAUACGUUUAUUUACAUGUGGGUAUGUAUGAGAGAAAUGAG .......((((((.(((((.(((((((..............((.(((((.((.......)).)))))))..(((((......)))))))))))).))))).))))))........... ( -27.90) >DroSim_CAF1 33762 118 - 1 GAUAAUACAUACACACCUGGAUGUGAAAUUGCCAAAAAACAGAACACAUACAAACAAGCUGCAUGUGUCAUUAAACCAAUACGUUUAUUUACAUGUGGGCAUGUAUGAGAGAAAUGAG .......((((((..((((.(((((((..............((.(((((.((.......)).)))))))..(((((......)))))))))))).))))..))))))........... ( -25.80) >DroEre_CAF1 40568 110 - 1 GAUAAUACAUACACACCUGGAUGUGAAACUGGCAAAAAACCGUACACAUACAAAUAAACUGCAUGUGUCAUUAAACCAAUACUUUUAUUUACAUGUG--------UGAGAGAAAUGAG ...........(((((.((...((((((..((.......))(.((((((.((.......)).))))))).............))))))...)).)))--------))........... ( -17.40) >consensus GAUAAUACAUACACACCUGGAUGUGAAAUUGCCAAAAAACAGAACACAUACAAACAAGCUGCAUGUGUCAUUAAACCAAUACGUUUAUUUACAUGUGGGUAUGUAUGAGAGAAAUGAG .......((((((.(((((.(((((((................((((((.((.......)).))))))...(((((......)))))))))))).))))).))))))........... (-20.12 = -22.38 + 2.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:54 2006