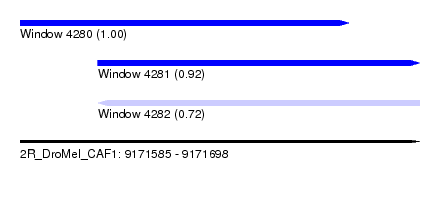
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,171,585 – 9,171,698 |
| Length | 113 |
| Max. P | 0.998448 |

| Location | 9,171,585 – 9,171,678 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -22.98 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9171585 93 + 20766785 ACAG------G--CACACACACUCACACAG--CGAACGGAACGGAACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGA ....------.--.........((((((.(--((((..........((....))....(((((((...(((.......)))..)))))))..))))))))))) ( -28.90) >DroSec_CAF1 55980 83 + 1 -------------CACACACACUCACACAG--CGAACGGA-----ACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGA -------------.........((((((.(--((((....-----.((....))....(((((((...(((.......)))..)))))))..))))))))))) ( -28.90) >DroSim_CAF1 48314 83 + 1 -------------CACACACACGCACACAG--CGAACGGA-----ACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGA -------------....(((((((......--........-----.((....))(((((((((((...(((.......)))..)))))).)))))))))))). ( -31.00) >DroEre_CAF1 52889 84 + 1 ACAC--A---GCA--------CACACAGAAC-GGAACGGA-----ACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGA ....--.---.((--------(((.(.....-........-----.((....))(((((((((((...(((.......)))..)))))).)))))).))))). ( -25.30) >DroAna_CAF1 49954 98 + 1 ACAGACACUCGCACACUCGCACACACAGAGGGAGAGUGAA-----ACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGA .....(((.(((.(((((...(.......)...)))))..-----.((....))(((((((((((...(((.......)))..)))))).)))))))).))). ( -32.80) >consensus ACA_______G__CACACACACACACACAG__CGAACGGA_____ACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGA .....................(((((....................((....))(((((((((((...(((.......)))..)))))).)))))..))))). (-22.98 = -23.30 + 0.32)

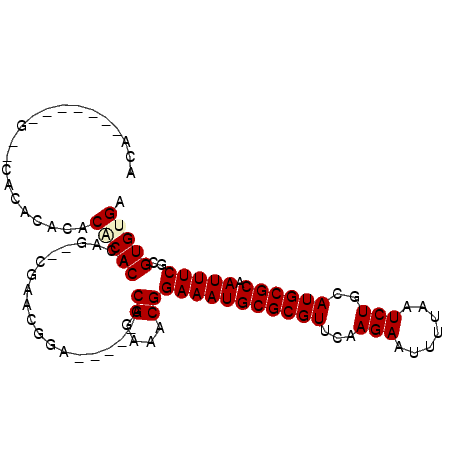
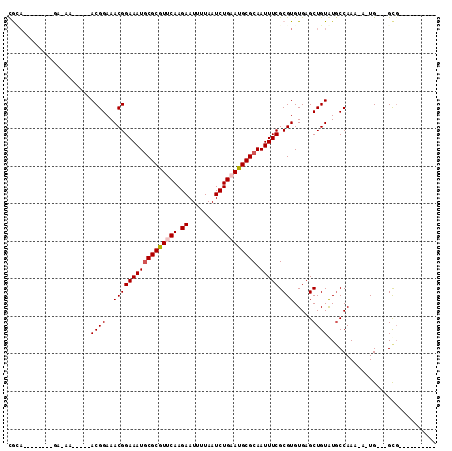
| Location | 9,171,607 – 9,171,698 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -21.64 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9171607 91 + 20766785 CGAA--------CG-GAACGGAACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAA-UG---GCG---------- ...(--------((-(.(((...((....))(((((((((((...(((.......)))..)))))).))))).))).....))))..((((....-))---)).---------- ( -28.00) >DroVir_CAF1 68869 86 + 1 CGCA--------GACAA-----ACGGAAACGGAAAUGCGCAUUCAAGAAUUUUAAUCUGAAUGCGCAAUUUCGCGUGUGAGCUGUGUUCCAAA-A-UG---GCG---------- (((.--------.....-----.((....))((((((((((((((.((.......))))))))))).)))))))).....(((((........-)-))---)).---------- ( -30.30) >DroPse_CAF1 40483 108 + 1 UGGAGGGAAAACUG-AA-----ACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGAAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAAUGUGGGCGCACACACACA ....(.((((....-..-----.((....))....((((((((((.((.......)))))))))))).)))).)(((((...(((.((((..........)))).)))))))). ( -35.50) >DroGri_CAF1 58670 84 + 1 CACU--------GA-AA-----ACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGAAUGCGCAAUUUCGCGUGUGAGCUGUGUGCCAAA-A-UG---GC----------- (((.--------..-..-----.((....))((((((((((((((.((.......))))))))))).)))))((......)).))).(((...-.-.)---))----------- ( -27.50) >DroWil_CAF1 71130 86 + 1 UAUA--------CA-AA-----ACGGAAACGGAAAUACGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAA-UG---GUG---------- ....--------..-..-----.((....))...(((((.((((.(((.......)))(((((((......))))))))))))))))((((....-))---)).---------- ( -22.50) >DroMoj_CAF1 73690 85 + 1 CGCA--------GA-AA-----ACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGAAUGCGCAAUUUCGCGUGUGAGCUGUGUGCCAAA-A-UG---GCG---------- ((((--------(.-..-----(((....)((((.((((((((((.((.......)))))))))))).))))....))...))))).(((...-.-.)---)).---------- ( -32.50) >consensus CGCA________GA_AA_____ACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGAAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAA_A_UG___GCG__________ ......................((((..(((((((((((((((((.((.......))))))))))).))))).))).....))))............................. (-21.64 = -22.00 + 0.36)

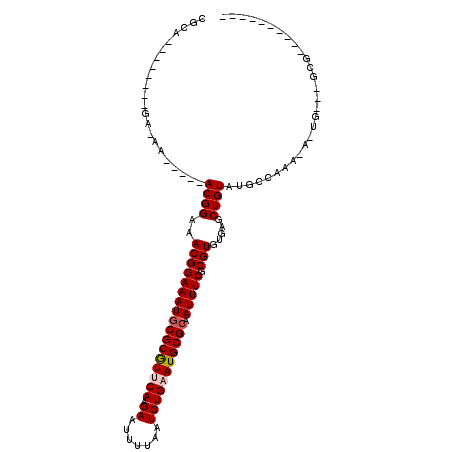
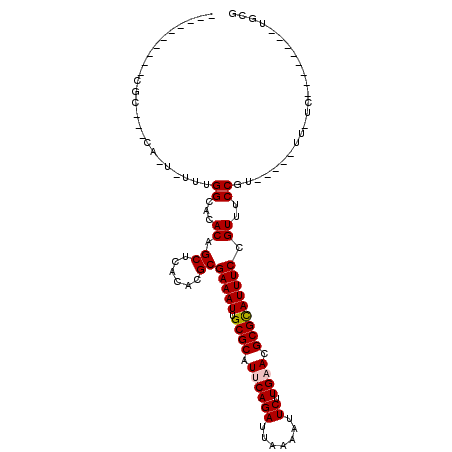
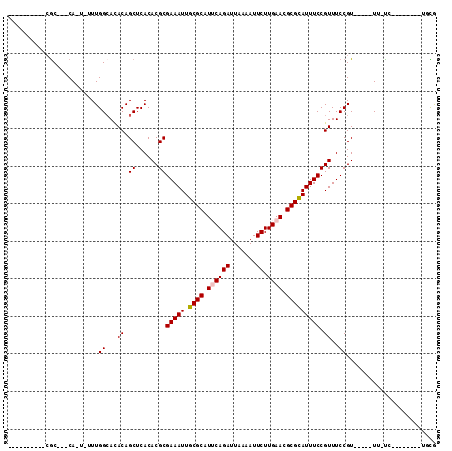
| Location | 9,171,607 – 9,171,698 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9171607 91 - 20766785 ----------CGC---CA-UUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUUCCGUUCCGUUC-CG--------UUCG ----------.((---((-....))))....(((....(((.(((((.((((...((((.......)).))...))))))))))))....)))......-..--------.... ( -20.30) >DroVir_CAF1 68869 86 - 1 ----------CGC---CA-U-UUUGGAACACAGCUCACACGCGAAAUUGCGCAUUCAGAUUAAAAUUCUUGAAUGCGCAUUUCCGUUUCCGU-----UUGUC--------UGCG ----------(((---((-.-..(((((.((.((......))(((((.(((((((((((.......)).)))))))))))))).))))))).-----.))..--------.))) ( -26.80) >DroPse_CAF1 40483 108 - 1 UGUGUGUGUGCGCCCACAUUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUUCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUUCCGU-----UU-CAGUUUUCCCUCCA .(((((((.(((((..........))).....)).)))))))(((((.((((.((((((.......)).)))).))))))))).........-----..-.............. ( -29.00) >DroGri_CAF1 58670 84 - 1 -----------GC---CA-U-UUUGGCACACAGCUCACACGCGAAAUUGCGCAUUCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUUCCGU-----UU-UC--------AGUG -----------((---(.-.-...))).((((((....(((.(((((.((((.((((((.......)).)))).))))))))))))....))-----).-..--------.))) ( -22.50) >DroWil_CAF1 71130 86 - 1 ----------CAC---CA-UUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGUAUUUCCGUUUCCGU-----UU-UG--------UAUA ----------..(---((-....))).((((((..(..(((.(((((.((((...((((.......)).))...))))))))))))....).-----.)-))--------))). ( -17.10) >DroMoj_CAF1 73690 85 - 1 ----------CGC---CA-U-UUUGGCACACAGCUCACACGCGAAAUUGCGCAUUCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUUCCGU-----UU-UC--------UGCG ----------.((---(.-.-...))).....((....(((.((((((((((.((((((.......)).)))).))))).....))))))))-----..-..--------.)). ( -23.50) >consensus __________CGC___CA_U_UUUGGCACACAGCUCACACGCGAAAUUGCGCAUUCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUUCCGU_____UU_UC________UGCG ........................((...((.((......))(((((.((((.((((((.......)).)))).))))))))).))..))........................ (-15.72 = -15.92 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:07 2006