
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,157,974 – 9,158,070 |
| Length | 96 |
| Max. P | 0.999819 |

| Location | 9,157,974 – 9,158,070 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.68 |
| SVM decision value | 4.16 |
| SVM RNA-class probability | 0.999819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
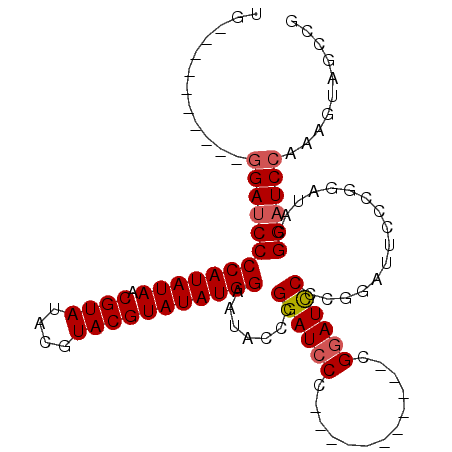
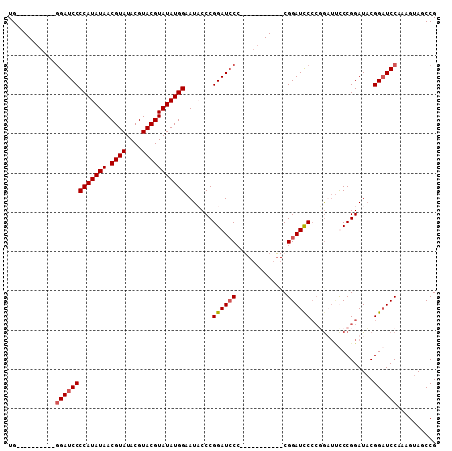
>2R_DroMel_CAF1 9157974 96 + 20766785 UG----------AGAUCCCCAUAUAACGUAUACGUACGUAUAUGGCAUCCCCGGAUCCCUGAAUC---CUCGGAUCCCCGAAUCCCCGGAUACGGUUCCAAAGUAGCCG ..----------.((((((((((((.((((....))))))))))).......((((......)))---)..))))).(((......)))...(((((.......))))) ( -26.60) >DroSec_CAF1 32219 88 + 1 UG----------GGAUCCCCAUAUAACGUAUACGUACGUAUAUGGAAUACCCGGAUCCC-----------CGGAUCCCCUGUUUCCCGGAUACGGAUCCAAAGUAGCCG .(----------(((((((((((((.((((....))))))))))).......)))))))-----------.((((((.(((.....)))....)))))).......... ( -31.61) >DroSim_CAF1 32134 88 + 1 UG----------GGAUCCCCAUAUAACGUAUACGUACGUAUAUGGAAUCCCCGGAUCCC-----------CGGAUCCCCUGUUUUCCGGAUACGGAUCCAAAGUAGCCG .(----------((((..(((((((.((((....))))))))))).))))).(((((((-----------((((..........)))))....)))))).......... ( -34.80) >DroEre_CAF1 35250 103 + 1 UGGUGCGAACUGGGAUCCCCAUAUAACGUAUACGUACGUAUAUGG-AUACCCGGAUGCG-GAUUCGUAUUCGGAUUC--GGA--UCCGGAUCCGGAUCCAAAGUAGCCG .(((....(((.(((((((((((((.((((....)))))))))))-....((((((.((-((((((....)))))))--).)--)))))....))))))..))).))). ( -47.90) >consensus UG__________GGAUCCCCAUAUAACGUAUACGUACGUAUAUGGAAUACCCGGAUCCC___________CGGAUCCCCGGAUUCCCGGAUACGGAUCCAAAGUAGCCG ............(((((((((((((.((((....))))))))))).......((((((.............))))))................)))))).......... (-23.98 = -24.54 + 0.56)

| Location | 9,157,974 – 9,158,070 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -26.00 |
| Energy contribution | -27.12 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.70 |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
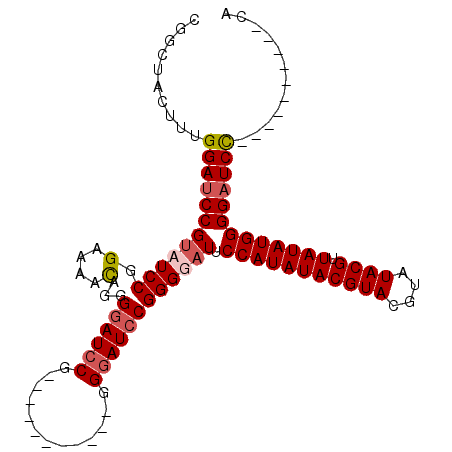
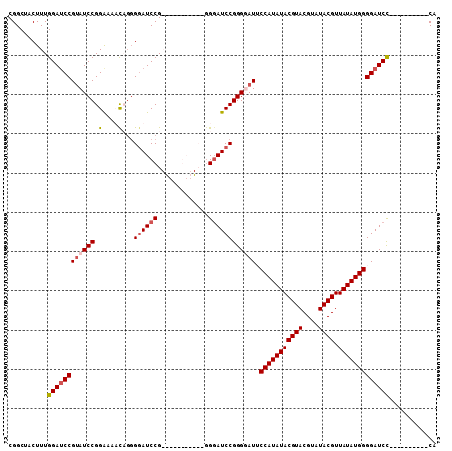
>2R_DroMel_CAF1 9157974 96 - 20766785 CGGCUACUUUGGAACCGUAUCCGGGGAUUCGGGGAUCCGAG---GAUUCAGGGAUCCGGGGAUGCCAUAUACGUACGUAUACGUUAUAUGGGGAUCU----------CA (((...(((((((......)))))))..)))(((((((..(---((((....))))).......(((((((((((....)))).)))))))))))))----------). ( -35.50) >DroSec_CAF1 32219 88 - 1 CGGCUACUUUGGAUCCGUAUCCGGGAAACAGGGGAUCCG-----------GGGAUCCGGGUAUUCCAUAUACGUACGUAUACGUUAUAUGGGGAUCC----------CA ..........((((((((((((.(....)...(((((..-----------..))))))))))).(((((((((((....)))).)))))))))))))----------.. ( -36.70) >DroSim_CAF1 32134 88 - 1 CGGCUACUUUGGAUCCGUAUCCGGAAAACAGGGGAUCCG-----------GGGAUCCGGGGAUUCCAUAUACGUACGUAUACGUUAUAUGGGGAUCC----------CA (((...((((((((((....((........)))))))))-----------)))..)))(((((((((((((((((....)))).))))).)))))))----------). ( -37.80) >DroEre_CAF1 35250 103 - 1 CGGCUACUUUGGAUCCGGAUCCGGA--UCC--GAAUCCGAAUACGAAUC-CGCAUCCGGGUAU-CCAUAUACGUACGUAUACGUUAUAUGGGGAUCCCAGUUCGCACCA .((....((((((((((....))))--)))--)))..(((((.((....-)).....(((..(-(((((((((((....)))).))))))))...))).)))))..)). ( -38.40) >consensus CGGCUACUUUGGAUCCGUAUCCGGAAAACAGGGGAUCCG___________GGGAUCCGGGGAUUCCAUAUACGUACGUAUACGUUAUAUGGGGAUCC__________CA ..........((((((((((((.(....)...((((((.............)))))))))))).(((((((((((....)))).)))))))))))))............ (-26.00 = -27.12 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:02 2006