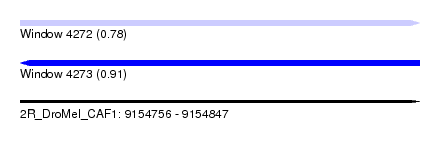
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,154,756 – 9,154,847 |
| Length | 91 |
| Max. P | 0.912855 |

| Location | 9,154,756 – 9,154,847 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
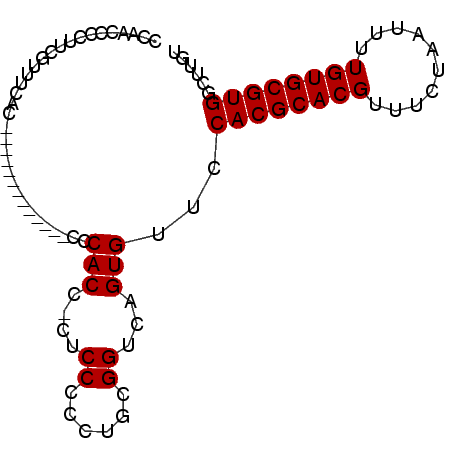
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -16.65 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9154756 91 + 20766785 ACAAGCCACGCACAAAAUUAGAAACGUGCGUGCAACACAGACCGCAGGAGGGGUGGUGGGGGGUAUGGUGUUGGUGAAACGAAGGGGUUGA .(((.((((((((............)))))).((((((..(((.(..............).)))...))))))...........)).))). ( -25.54) >DroSec_CAF1 29124 86 + 1 ACAAGCCACGCACAAAAUUAGAAACGUGCGUGCAACACAGACCGCAGGGGGAGGGGUGGGGG---CGGUGU--GUGAAACGAAGGGGUUGG .(((.((((((((............))))))....((((.(((((................)---)))).)--)))........)).))). ( -27.89) >DroEre_CAF1 32247 77 + 1 ACAAGCCACGCACAAAAUUAGAAACGUGCGUGGAACACUGACCGCAGGGGGAG-GGUGGG-------------GUGAACCGAAGGGGUAGG .....((((((((............))))))))..((((..((((........-.)))))-------------)))..((....))..... ( -24.30) >DroYak_CAF1 30067 77 + 1 ACAAGCCACGCACAAAAUUAGAAACGUGCGUGGAACACUGACCGCAGGGGGUG-GGUGGG-------------GUGUAUCGAAGGGGUUGG (((..((((((((............))))))))..((((.(((......))).-))))..-------------.))).((....))..... ( -25.10) >consensus ACAAGCCACGCACAAAAUUAGAAACGUGCGUGCAACACAGACCGCAGGGGGAG_GGUGGG_____________GUGAAACGAAGGGGUUGG .(((.((((((((............))))))..........((((..........)))).........................)).))). (-16.65 = -16.90 + 0.25)

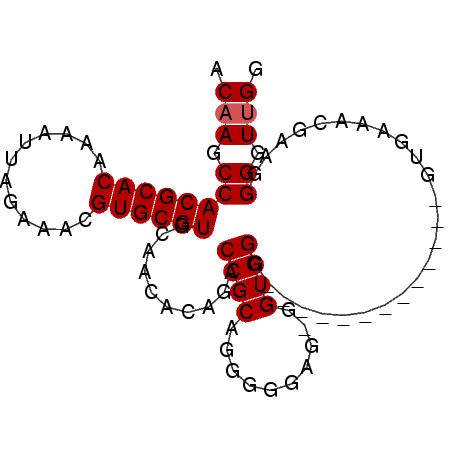
| Location | 9,154,756 – 9,154,847 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -20.14 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9154756 91 - 20766785 UCAACCCCUUCGUUUCACCAACACCAUACCCCCCACCACCCCUCCUGCGGUCUGUGUUGCACGCACGUUUCUAAUUUUGUGCGUGGCUUGU ..................((((((...(((..................)))..))))))((((((((..........))))))))...... ( -18.37) >DroSec_CAF1 29124 86 - 1 CCAACCCCUUCGUUUCAC--ACACCG---CCCCCACCCCUCCCCCUGCGGUCUGUGUUGCACGCACGUUUCUAAUUUUGUGCGUGGCUUGU .(((.((....((..(((--(.((((---(................))))).))))..))(((((((..........))))))))).))). ( -25.09) >DroEre_CAF1 32247 77 - 1 CCUACCCCUUCGGUUCAC-------------CCCACC-CUCCCCCUGCGGUCAGUGUUCCACGCACGUUUCUAAUUUUGUGCGUGGCUUGU ...(((.....))).(((-------------...(((-(.......).)))..)))..(((((((((..........)))))))))..... ( -17.90) >DroYak_CAF1 30067 77 - 1 CCAACCCCUUCGAUACAC-------------CCCACC-CACCCCCUGCGGUCAGUGUUCCACGCACGUUUCUAAUUUUGUGCGUGGCUUGU ..............((((-------------...(((-((.....)).)))..)))).(((((((((..........)))))))))..... ( -19.20) >consensus CCAACCCCUUCGUUUCAC_____________CCCACC_CUCCCCCUGCGGUCAGUGUUCCACGCACGUUUCUAAUUUUGUGCGUGGCUUGU .................................(((....((......))...)))...((((((((..........))))))))...... (-13.73 = -13.72 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:59 2006