
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,130,844 – 9,130,935 |
| Length | 91 |
| Max. P | 0.500000 |

| Location | 9,130,844 – 9,130,935 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -15.10 |
| Consensus MFE | -8.57 |
| Energy contribution | -8.52 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
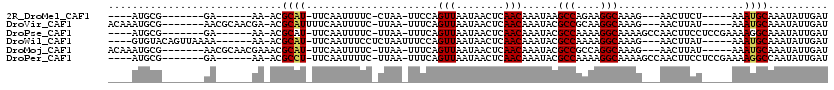
>2R_DroMel_CAF1 9130844 91 - 20766785 ----AUGCG-------GA------AA-ACGCAU-UUCAAUUUUC-CUAA-UUCCAGUUAAUAACUCAACAAAUAAGCCAGAAGGCAAAG---AACUUCU-----AAAUGCAAAUAUUGAU ----(((((-------..------..-.)))))-.(((((....-....-.....(((........)))......((.(((((......---..)))))-----....))....))))). ( -12.70) >DroVir_CAF1 11440 102 - 1 ACAAAUGCG-------AACGCAACGA-ACGCAUUUUCAAUUUUC-UUAA-UUUCAGUUAAUAACUCAACAAAUACGCCGCAAGGCAAAG---AACUUAU-----AAAUGCAAAUAUUGAU .(((.(((.-------...)))....-..((((((......(((-((..-.....(((........)))......(((....))).)))---)).....-----)))))).....))).. ( -19.90) >DroPse_CAF1 4918 99 - 1 ----AUGCG-------GA------AA-ACGCAU-UUCAAUUUUC-UUAA-UUUCAGUUAAUAACUCAACAAAUACGCCAAAAGGCAAAAGCCAACUUCCUCCGAAAAGGCAAAUAUUGAU ----(((((-------..------..-.)))))-..........-....-.....(((((((.............(((....)))....(((...(((....)))..)))...))))))) ( -17.00) >DroWil_CAF1 8678 100 - 1 ----GUGUACAGUUAAAA------AA-ACGCAU-UUCAAUUUCCUCUAAUUUCCAGUUAAUAACUCAACAAAUACGCCAAAAGGCAAAG---AACUUAU-----AAAUGCAAAUAUUGAU ----..............------..-..((((-((....(((............(((........)))......(((....)))...)---)).....-----)))))).......... ( -10.10) >DroMoj_CAF1 7402 102 - 1 ACAAAUGCG-------AACGCAACGAAACGCAU-UUCAAUUUUC-UUAA-UUUCAGUUAAUAACUCAACAAAUACGCCGCCAGGCAAAG---AACUUAU-----AAAUGCAAAUAUUGAU .(((.(((.-------...))).......((((-((.....(((-((..-.....(((........)))......(((....))).)))---)).....-----)))))).....))).. ( -16.80) >DroPer_CAF1 5164 99 - 1 ----AUGCG-------GA------AA-ACGCCU-UUCAAUUUUC-UUAA-UUUCAGUUAAUAACUCAACAAAUACGCCAAAAGGCAAAAGCCAACUUCCUCCGAAAAGGCCAAUAUUGAU ----..(((-------..------..-.)))..-..........-....-.....(((((((.............(((....)))....(((...(((....)))..)))...))))))) ( -14.10) >consensus ____AUGCG_______AA______AA_ACGCAU_UUCAAUUUUC_UUAA_UUUCAGUUAAUAACUCAACAAAUACGCCAAAAGGCAAAG___AACUUAU_____AAAUGCAAAUAUUGAU .............................((((......................(((........)))......(((....))).....................)))).......... ( -8.57 = -8.52 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:51 2006