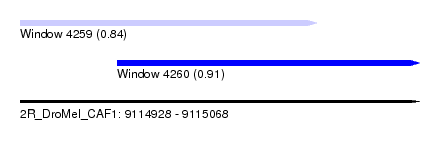
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,114,928 – 9,115,068 |
| Length | 140 |
| Max. P | 0.910355 |

| Location | 9,114,928 – 9,115,032 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.35 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -15.87 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9114928 104 + 20766785 AAUCUUGCUUUUGGCCCUUUUUCUUGUUGUUGCA---G-----CACUGUUA-CGAGGGCGGCUUAUUAGCCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGCAAAAGGG-GCCA ...........((((((((((((((((....(((---.-----...))).)-)))).((((((((((((((.((((......))))))))..)))))))))))))))))-)))) ( -41.20) >DroPse_CAF1 4526 104 + 1 UGUCGGUCUUUUGGCCUU------UGUUGGGGCA---GCAGAGCCCUUUUG-CGAGGCCAGCUUAUUAGGCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGAAAAACGGUGCCA ....(((....(((((((------.((.(((((.---.....)))))...)-))))))))((((((((((((((((......))))))).)))))))))...........))). ( -39.00) >DroEre_CAF1 3174 104 + 1 AAUCUGGCUUUUGGCCCUUUUUCUUGUUGCUGCA---G-----CACUGCUG-CGAGGGCGGCUUAUUAGCCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGAAAAAGGG-GCCA ...........((((((((((((..(((..((((---(-----(...))))-))..)))((((((((((((.((((......))))))))..)))))))))))))).))-)))) ( -40.30) >DroYak_CAF1 3188 103 + 1 AAUCUUGCUUUUGGGCCUUU-UCCUGUUGUUGCU---G-----CACUGUUA-CGAGGGCGGCUUAUUAGCCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGAAAAAGGG-GCCA .............(((((((-(.((.((((.((.---.-----....)).)-))).))(((((((((((((.((((......))))))))..)))))))))...)))))-))). ( -32.90) >DroAna_CAF1 3184 103 + 1 --UCCGCCCUCUGUCUCACUUUCUUUUUGGCCUG---G-----CCCUUUUGUCGAGGGCGGCUUAUUAGCCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGAAAAACGG-UGCC --..........................((((((---(-----(((((.....))))))((((((((((((.((((......))))))))..))))))))......)))-.))) ( -31.10) >DroPer_CAF1 4668 107 + 1 UCUCGGUCUUGUGGCCUU------UGUUGGGGCAGCAGCAGAGCCCUUAUG-CGAGGCCAGCUUAUUAGGCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGAAAAACGGUGCCA ....(((.((((((((((------.((.(((((.........)))))...)-))))))).((((((((((((((((......))))))).)))))))))......)))).))). ( -40.20) >consensus AAUCUGGCUUUUGGCCCUUU_UCUUGUUGGUGCA___G_____CACUGUUG_CGAGGGCGGCUUAUUAGCCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGAAAAACGG_GCCA ............(((((............................((........)).(((((((((((.((((((......))))))..)))))))))))......)).))). (-15.87 = -15.90 + 0.03)

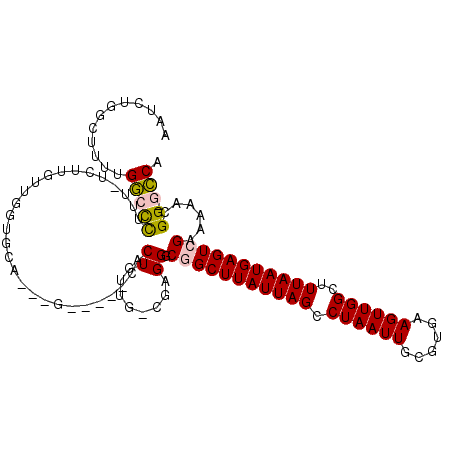

| Location | 9,114,962 – 9,115,068 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -21.51 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9114962 106 + 20766785 G-----CACUGUUA-CGAGGGCGGCUUAUUAGCCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGCAAAAGGG-GCCAAGCCUGGCCC----ACAAAAGGCCACAAAGCAAAAGGCGC (-----(..((((.-.(.((((((((((((((((.((((......))))))))..)))))))))).....((-((((....))))))----........)).)..))))....)).. ( -37.20) >DroVir_CAF1 5873 104 + 1 G-----C-CUACUU-CU-GGGCGGCUUAUUAGGCUAAUUGCGUAAAGUUGGCUUUAAUGAGUCGAAAAACGGUGCCAAGCCUGGC-C----ACAAAAAGCCACAAAGCGUGCGGCAA (-----(-((((((-.(-(((((((((((((((((((((......))))))).))))))))))........((((((....))).-)----)).....))).)).)).))).))).. ( -36.60) >DroPse_CAF1 4554 103 + 1 GCAGAGCCCUUUUG-CGAGGCCAGCUUAUUAGGCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGAAAAACGGUGCCAAGUCGGGCCC----GCAAAAGGCCACAAAGG--------- .....(.(((((((-((.((((.((((((((((((((((......))))))).)))))))))(((.....(....)...))))))))----)))))))).).......--------- ( -39.10) >DroGri_CAF1 3600 98 + 1 G-----C-CUACUU-CG-GGGCGGCUUAUUAGGCUAAUUGCGUAAAGUUGGCUUUAAUGAGUCGAAAAACGGUGCCAAGCCUGGC-C----ACAAAAAGCCACAAAGCA------AA (-----(-...(..-..-)((((((((((((((((((((......))))))).))))))))))........((((((....))).-)----)).....))).....)).------.. ( -32.10) >DroMoj_CAF1 5155 104 + 1 G-----C-CUACUU-CG-GGGCGGCUUAUUAGGCUAAUUGCGUAAAGUUGGCUUUAAUGAGUCGAAAAACGGUGCCAAGCCUGGC-C----ACAAAAAGCCACAAAGCAAGCGCCAA (-----(-((....-..-))))(((((((((((((((((......))))))).)))))))))).......(((((...((.((((-.----.......))))....))..))))).. ( -37.60) >DroAna_CAF1 3216 111 + 1 G-----CCCUUUUGUCGAGGGCGGCUUAUUAGCCUAAUUGCGUGAAGUUGGCUUUAAUGAGUCGAAAAACGG-UGCCAGCCAGGGCCCGGGCCAGAAAGCCACAAAGCAAAAGUGGC (-----(((((((((...((((((((((((((((.((((......))))))))..)))))))).......((-(....)))...)))).(((......))).....))))))).))) ( -41.50) >consensus G_____C_CUACUU_CG_GGGCGGCUUAUUAGGCUAAUUGCGUAAAGUUGGCUUUAAUGAGUCGAAAAACGGUGCCAAGCCUGGC_C____ACAAAAAGCCACAAAGCAA__G_CAA ...................((((((((((((((((((((......))))))).))))))))))..........(((......))).............)))................ (-21.51 = -22.07 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:46 2006