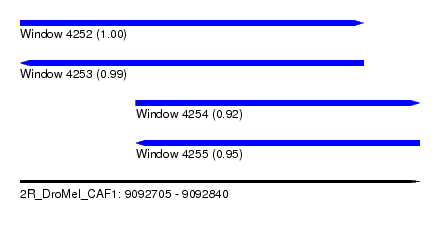
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,092,705 – 9,092,840 |
| Length | 135 |
| Max. P | 0.997529 |

| Location | 9,092,705 – 9,092,821 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -24.91 |
| Energy contribution | -24.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
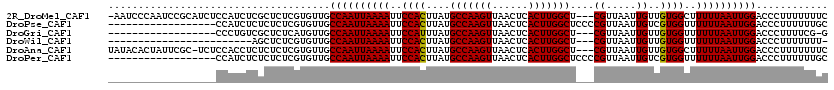
>2R_DroMel_CAF1 9092705 116 + 20766785 -AAUCCCAAUCCGCAUCUCCAUCUCGCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCU---CGUUAAUUGUUGUGGCUUUUUAAUUGGACCCUUUUUUUC -..................((...(((....))).))((((((((((..((((....(((((((......))))))).---((.....))..))))..))))))))))............ ( -25.00) >DroPse_CAF1 2338 102 + 1 ------------------CCAUCUCUCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUCCCCGUUAAUUGUCGUGGUUUUUUAAUUGGACCCUUUUUUGC ------------------...................((((((((((..((((....(((((((......))))))).....(........)))))..))))))))))............ ( -25.90) >DroGri_CAF1 3159 98 + 1 ------------------CCCUGUCGCUCUCAUGUUGCCAAUUAAAAUUCCAUUUAUGCCAAGUUAACUCACUUGGCU---CGUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUCG-G ------------------...................((((((((((..((((....(((((((......))))))).---...........))))..))))))))))..........-. ( -23.79) >DroWil_CAF1 4013 92 + 1 ------------------------AGCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCU---CGUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUUUU- ------------------------.............((((((((((..((((....(((((((......))))))).---((.....))..))))..))))))))))...........- ( -25.30) >DroAna_CAF1 3104 116 + 1 UAUACACUAUUCGC-UCUCCACCUCUCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCU---CGUUAAUUGUUGUGGCUUUUUAAUUGGACCCUUUUUUUC ...((((.......-................))))..((((((((((..((((....(((((((......))))))).---((.....))..))))..))))))))))............ ( -24.80) >DroPer_CAF1 3887 102 + 1 ------------------CCAUCUCUCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUCCCCGUUAAUUGUCGUGGUUUUUUAAUUGGACCCUUUUUUGC ------------------...................((((((((((..((((....(((((((......))))))).....(........)))))..))))))))))............ ( -25.90) >consensus __________________CCAUCUCGCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCU___CGUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUUUUC .....................................((((((((((..((((....(((((((......)))))))....((.....))..))))..))))))))))............ (-24.91 = -24.77 + -0.14)
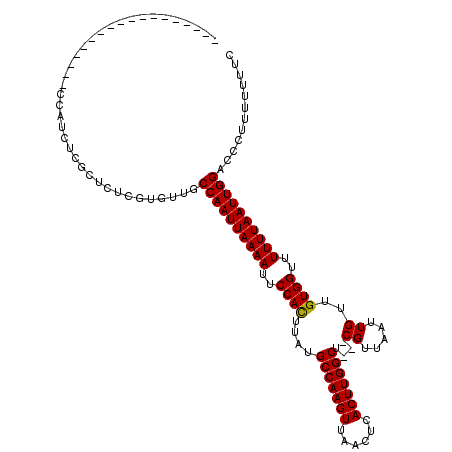
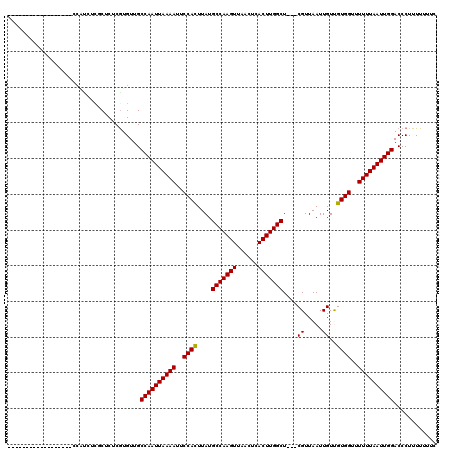
| Location | 9,092,705 – 9,092,821 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -24.37 |
| Energy contribution | -24.53 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9092705 116 - 20766785 GAAAAAAAGGGUCCAAUUAAAAAGCCACAACAAUUAACG---AGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGCGAGAUGGAGAUGCGGAUUGGGAUU- ............((((((((((..((((..(.......)---.(((((((......)))))))....))))..))))))))))(((..((....(........)...))..))).....- ( -29.20) >DroPse_CAF1 2338 102 - 1 GCAAAAAAGGGUCCAAUUAAAAAACCACGACAAUUAACGGGGAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGAGAGAUGG------------------ ..........(.((((((((((..((((..(........)...(((((((......)))))))....))))..)))))))))))....(....)........------------------ ( -26.10) >DroGri_CAF1 3159 98 - 1 C-CGAAAAGGGUCCAAUUAAAAAACCACAACAAUUAACG---AGCCAAGUGAGUUAACUUGGCAUAAAUGGAAUUUUAAUUGGCAACAUGAGAGCGACAGGG------------------ (-(.....))(.((((((((((..(((...(.......)---.(((((((......))))))).....)))..)))))))))))..................------------------ ( -22.90) >DroWil_CAF1 4013 92 - 1 -AAAAAAAGGGUCCAAUUAAAAAACCACAACAAUUAACG---AGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGCU------------------------ -.........(.((((((((((..((((..(.......)---.(((((((......)))))))....))))..)))))))))))............------------------------ ( -25.50) >DroAna_CAF1 3104 116 - 1 GAAAAAAAGGGUCCAAUUAAAAAGCCACAACAAUUAACG---AGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGAGAGGUGGAGA-GCGAAUAGUGUAUA ..........(.((((((((((..((((..(.......)---.(((((((......)))))))....))))..))))))))))).((((................-.......))))... ( -28.70) >DroPer_CAF1 3887 102 - 1 GCAAAAAAGGGUCCAAUUAAAAAACCACGACAAUUAACGGGGAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGAGAGAUGG------------------ ..........(.((((((((((..((((..(........)...(((((((......)))))))....))))..)))))))))))....(....)........------------------ ( -26.10) >consensus GAAAAAAAGGGUCCAAUUAAAAAACCACAACAAUUAACG___AGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGAGAGAUGG__________________ ..........(.((((((((((..((((..(.......)....(((((((......)))))))....))))..))))))))))).................................... (-24.37 = -24.53 + 0.17)

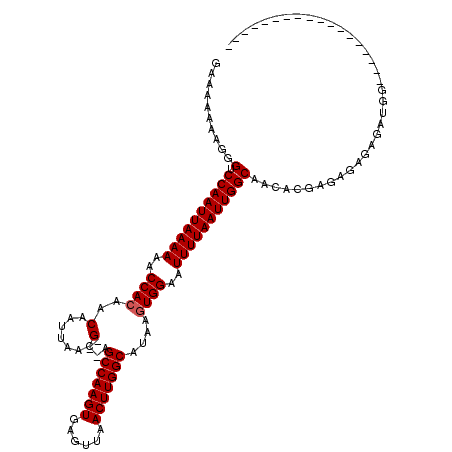
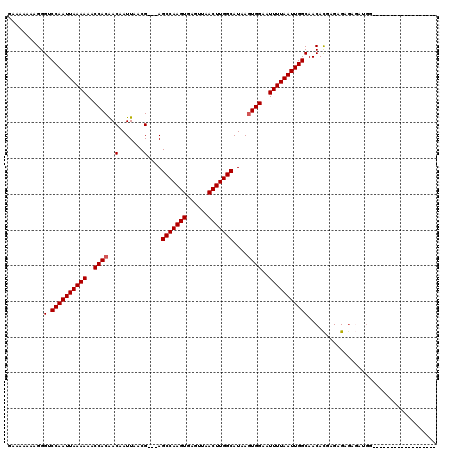
| Location | 9,092,744 – 9,092,840 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -19.26 |
| Consensus MFE | -16.52 |
| Energy contribution | -16.27 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9092744 96 + 20766785 AUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUCGUUAAUUGUUGUGGCUUUUUAAUUGGACCCUUUUUUUCUCCUCCUGCUG-----------GCUGCUUU (((((((..((((....(((((((......))))))).((.....))..))))..))))))).(((...........)))....((..-----------...))... ( -18.00) >DroVir_CAF1 2824 84 + 1 AUUAAAAUUCCAUUUAUGCCAAGUUAACUCACUUGGCUCGUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUCG-GAG---CUGAUG------------------- ..(((((..((((....(((((((......)))))))............))))..)))))((..(..((.....)-)..---)..)).------------------- ( -19.09) >DroGri_CAF1 3181 102 + 1 AUUAAAAUUCCAUUUAUGCCAAGUUAACUCACUUGGCUCGUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUCG-GUG---CUGGUGGUGCUGCGGCUGCUGCUC- (((((((..((((....(((((((......)))))))............))))..))))))).(.(((......)-)).---).((..((((....)).))..)).- ( -24.99) >DroWil_CAF1 4029 76 + 1 AUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUCGUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUUUU------------------------------- (((((((..((((....(((((((......))))))).((.....))..))))..)))))))..............------------------------------- ( -17.30) >DroMoj_CAF1 3171 84 + 1 AUUAAAAUUCCAUUUAUGCCAAGUUAACUCACUUGGCUCGUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUCG-GUG---CUGCUG------------------- (((((((..((((....(((((((......)))))))............))))..))))))).(.(((......)-)).---).....------------------- ( -18.99) >DroAna_CAF1 3143 91 + 1 AUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUCGUUAAUUGUUGUGGCUUUUUAAUUGGACCCUUUUUUUCUCCUGCGCCCA-----------ACU----- .((((((..((((....(((((((......))))))).((.....))..))))..))))))((((.(.(............).).)))-----------)..----- ( -17.20) >consensus AUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUCGUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUCG_GUG___CUGCUG___________________ (((((((..((((....(((((((......)))))))............))))..)))))))............................................. (-16.52 = -16.27 + -0.25)

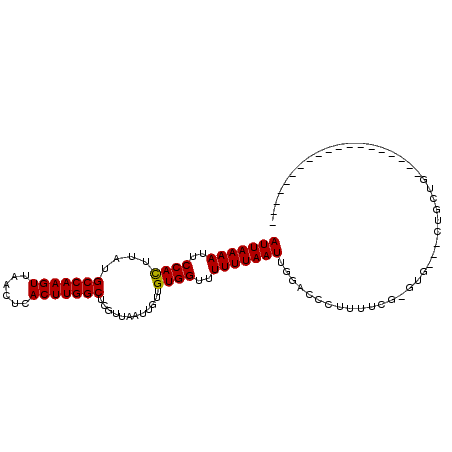
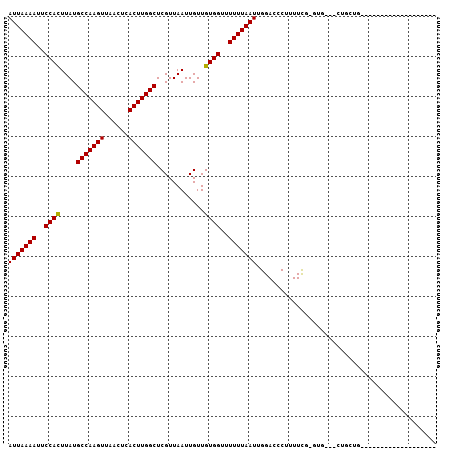
| Location | 9,092,744 – 9,092,840 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -14.75 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9092744 96 - 20766785 AAAGCAGC-----------CAGCAGGAGGAGAAAAAAAGGGUCCAAUUAAAAAGCCACAACAAUUAACGAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAU ...((...-----------..))....(((...........))).(((((((..((((..(.......).(((((((......)))))))....))))..))))))) ( -21.50) >DroVir_CAF1 2824 84 - 1 -------------------CAUCAG---CUC-CGAAAAGGGUCCAAUUAAAAAACCACAACAAUUAACGAGCCAAGUGAGUUAACUUGGCAUAAAUGGAAUUUUAAU -------------------......---.((-((.....(((...........)))..............(((((((......))))))).....))))........ ( -16.20) >DroGri_CAF1 3181 102 - 1 -GAGCAGCAGCCGCAGCACCACCAG---CAC-CGAAAAGGGUCCAAUUAAAAAACCACAACAAUUAACGAGCCAAGUGAGUUAACUUGGCAUAAAUGGAAUUUUAAU -..((.((....)).))....((((---.((-(......))).)..........................(((((((......))))))).....)))......... ( -20.90) >DroWil_CAF1 4029 76 - 1 -------------------------------AAAAAAAGGGUCCAAUUAAAAAACCACAACAAUUAACGAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAU -------------------------------..............(((((((..((((..(.......).(((((((......)))))))....))))..))))))) ( -17.70) >DroMoj_CAF1 3171 84 - 1 -------------------CAGCAG---CAC-CGAAAAGGGUCCAAUUAAAAAACCACAACAAUUAACGAGCCAAGUGAGUUAACUUGGCAUAAAUGGAAUUUUAAU -------------------.....(---.((-(......))).).(((((((..(((...(.......).(((((((......))))))).....)))..))))))) ( -16.40) >DroAna_CAF1 3143 91 - 1 -----AGU-----------UGGGCGCAGGAGAAAAAAAGGGUCCAAUUAAAAAGCCACAACAAUUAACGAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAU -----.((-----------(((((.(............).)))))))(((((..((((..(.......).(((((((......)))))))....))))..))))).. ( -25.90) >consensus ___________________CAGCAG___CAC_AAAAAAGGGUCCAAUUAAAAAACCACAACAAUUAACGAGCCAAGUGAGUUAACUUGGCAUAAAUGGAAUUUUAAU ..................................((((...((((.(((...........(.......).(((((((......))))))).))).)))).))))... (-14.75 = -14.75 + 0.00)

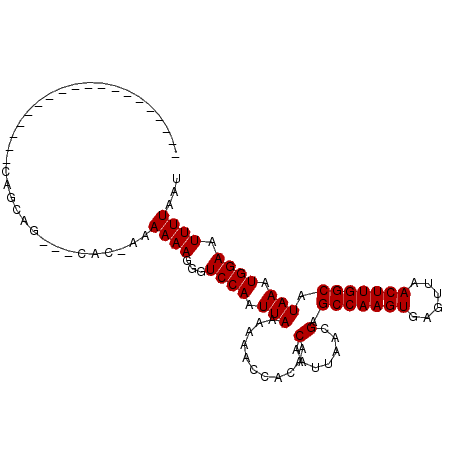
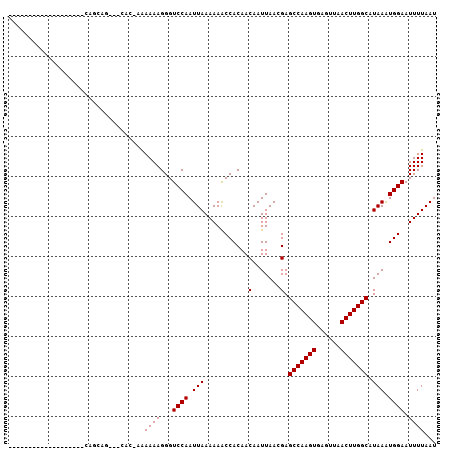
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:41 2006