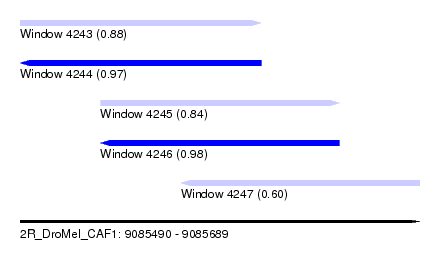
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,085,490 – 9,085,689 |
| Length | 199 |
| Max. P | 0.975822 |

| Location | 9,085,490 – 9,085,610 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -25.23 |
| Energy contribution | -24.98 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9085490 120 + 20766785 GCUGUUUUUUAUGUUUGCCUGGUGCAUGAAUAUUGAUUUUUCUUGAUUAAGCUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGUCCAGGCUGCAUUACAAAUGUAUUUA ...((((...((((..(((((.((((.(....((((((......)))))).)))))..............((((((((..........))))))))))))).))))...))))....... ( -27.60) >DroSec_CAF1 50264 119 + 1 GCUGUU-UUUAUGUUUGCCUGGUGCAUGAAUAUUGAUUUUUCUUGAUUAAGCUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGCCCAGGCUGCAUUUCAAAUGUAUUUU ...(((-(..((((..(((((.((((.(....((((((......)))))).)))))..............((((((((..........))))))))))))).))))...))))....... ( -30.00) >DroSim_CAF1 52476 120 + 1 GCUGUUUUUUAUGUUUUCUUGGUGCAUGAAUAUUAAUUUUUCUUGAUUAAGCUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGCCCAGGCUGCAUUACAAAUGUAUUUA ...((((...((((...((((.((((.(....((((((......)))))).)))))..............((((((((..........))))))))))))..))))...))))....... ( -23.80) >DroEre_CAF1 57589 120 + 1 GCUGUUAUUUUUGCUUGCCUGGUGCAUGAAAAUGGAUUUUUCUUGAUUAAACUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGCCCAGGCUGCAUUAGAAAAAUAUUUA ..((((.(((((((..((((((((..((((((((.(.(((........))).).).))))))))))....((((((((..........))))))))))))).)))..)))).)))).... ( -30.80) >consensus GCUGUUUUUUAUGUUUGCCUGGUGCAUGAAUAUUGAUUUUUCUUGAUUAAGCUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGCCCAGGCUGCAUUACAAAUGUAUUUA ...........(((..(((((.((((.(....((((((......)))))).)))))..............((((((((..........))))))))))))).)))............... (-25.23 = -24.98 + -0.25)

| Location | 9,085,490 – 9,085,610 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -27.75 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9085490 120 - 20766785 UAAAUACAUUUGUAAUGCAGCCUGGACAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGCUUAAUCAAGAAAAAUCAAUAUUCAUGCACCAGGCAAACAUAAAAAACAGC ....(((....))).....((((((.((((((((......))))))))))(((.(((((((.((((...(((....)))......)))).))))))))))))))................ ( -31.10) >DroSec_CAF1 50264 119 - 1 AAAAUACAUUUGAAAUGCAGCCUGGGCAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGCUUAAUCAAGAAAAAUCAAUAUUCAUGCACCAGGCAAACAUAAA-AACAGC ...................((((.((((((((((......))))))))))(((.(((((((.((((...(((....)))......)))).)))))))))))))).........-...... ( -34.60) >DroSim_CAF1 52476 120 - 1 UAAAUACAUUUGUAAUGCAGCCUGGGCAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGCUUAAUCAAGAAAAAUUAAUAUUCAUGCACCAAGAAAACAUAAAAAACAGC ....(((....))).......(((((((((((((......))))))))))(((.(((((((.((((...(((....)))......)))).))))))))))................))). ( -28.60) >DroEre_CAF1 57589 120 - 1 UAAAUAUUUUUCUAAUGCAGCCUGGGCAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGUUUAAUCAAGAAAAAUCCAUUUUCAUGCACCAGGCAAGCAAAAAUAACAGC ....((((((.....(((.((((.((((((((((......))))))))))(((.((((((((((.(...(((......)))....).)))))))))))))))))..)))))))))..... ( -36.00) >consensus UAAAUACAUUUGUAAUGCAGCCUGGGCAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGCUUAAUCAAGAAAAAUCAAUAUUCAUGCACCAGGCAAACAUAAAAAACAGC ...................((((.((((((((((......))))))))))(((.(((((((.((((...(((....)))......)))).))))))))))))))................ (-27.75 = -28.38 + 0.62)

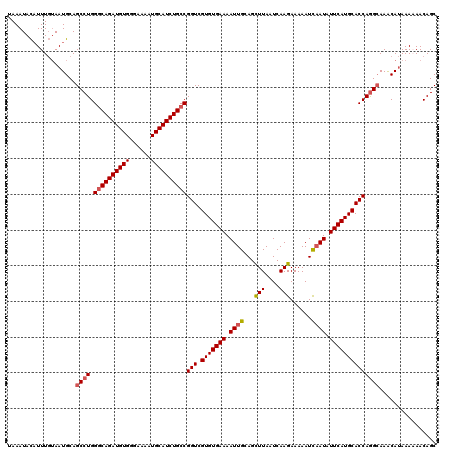
| Location | 9,085,530 – 9,085,649 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.66 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9085530 119 + 20766785 UCUUGAUUAAGCUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGUCCAGGCUGCAUUACAAAUGUAUUUAAAAUGCAUUUAUUUUUAUUCCCAUGCAACAGGU-UGCAGG ...........(((((((((......(.((((((((((..........)))))).)).)))(((((...((((((((.....))))))))............)))))..))))-))))). ( -26.26) >DroSec_CAF1 50303 119 + 1 UCUUGAUUAAGCUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGCCCAGGCUGCAUUUCAAAUGUAUUUUAAAUGCAUUUAUUUUCUUUCCCAUGCAACAGGU-UGCAGG ...........(((((((((......(.((((((((((..........))))))))..)))(((((...((((((((.....))))))))............)))))..))))-))))). ( -28.66) >DroSim_CAF1 52516 119 + 1 UCUUGAUUAAGCUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGCCCAGGCUGCAUUACAAAUGUAUUUAAAAUGCAUUUAUUUUCUUUCCCAUGCAACAGGU-UGCAGG ...........(((((((((......(.((((((((((..........))))))))..)))(((((...((((((((.....))))))))............)))))..))))-))))). ( -28.66) >DroEre_CAF1 57629 120 + 1 UCUUGAUUAAACUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGCCCAGGCUGCAUUAGAAAAAUAUUUAAAAUGCAUUUAUUUUCUUUGCCAUGCAACAGGUUUGCAGG ...........(((((..........((((((((((((..........))))))))..(((((((((..(((....)))..))))))............)))........))))))))). ( -29.30) >consensus UCUUGAUUAAGCUGCAAUUUUCACACGACCGGCAGAUGCAUUUUCCCACAUCUGCCCAGGCUGCAUUACAAAUGUAUUUAAAAUGCAUUUAUUUUCUUUCCCAUGCAACAGGU_UGCAGG ...........(((((...........(((((((((((..........))))))))...(.(((((...((((((((.....))))))))............))))).).))).))))). (-24.35 = -24.66 + 0.31)

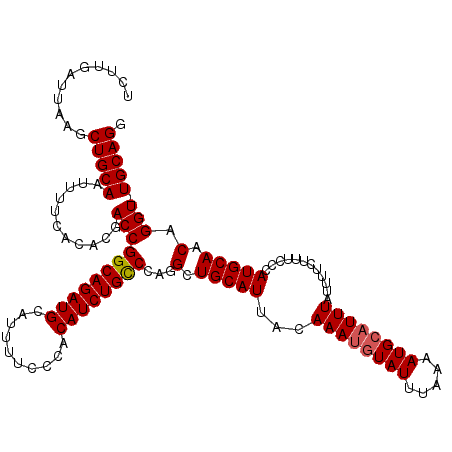
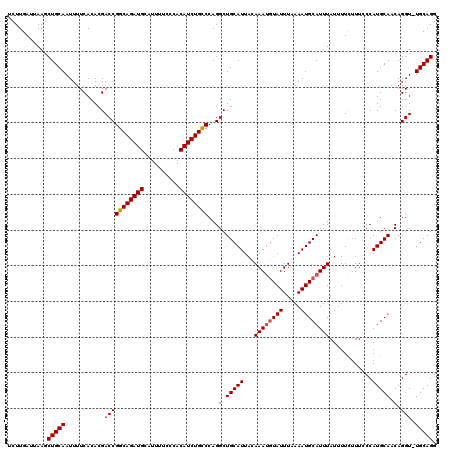
| Location | 9,085,530 – 9,085,649 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -33.30 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9085530 119 - 20766785 CCUGCA-ACCUGUUGCAUGGGAAUAAAAAUAAAUGCAUUUUAAAUACAUUUGUAAUGCAGCCUGGACAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGCUUAAUCAAGA .(((((-(....(..(((((.............((((((.((((....)))).))))))..((((.((((((((......)))))))))))))))))..)...))))))........... ( -35.30) >DroSec_CAF1 50303 119 - 1 CCUGCA-ACCUGUUGCAUGGGAAAGAAAAUAAAUGCAUUUAAAAUACAUUUGAAAUGCAGCCUGGGCAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGCUUAAUCAAGA .(((((-(....(..((((..............(((((((.(((....))).)))))))(((..((((((((((......)))))))))))))))))..)...))))))........... ( -36.90) >DroSim_CAF1 52516 119 - 1 CCUGCA-ACCUGUUGCAUGGGAAAGAAAAUAAAUGCAUUUUAAAUACAUUUGUAAUGCAGCCUGGGCAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGCUUAAUCAAGA .(((((-(....(..((((..............((((((.((((....)))).))))))(((..((((((((((......)))))))))))))))))..)...))))))........... ( -37.00) >DroEre_CAF1 57629 120 - 1 CCUGCAAACCUGUUGCAUGGCAAAGAAAAUAAAUGCAUUUUAAAUAUUUUUCUAAUGCAGCCUGGGCAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGUUUAAUCAAGA .((((((.....(..((((((............((((((..(((....)))..)))))).....((((((((((......)))))))))).))))))..)...))))))........... ( -39.60) >consensus CCUGCA_ACCUGUUGCAUGGGAAAGAAAAUAAAUGCAUUUUAAAUACAUUUGUAAUGCAGCCUGGGCAGAUGUGGGAAAAUGCAUCUGCCGGUCGUGUGAAAAUUGCAGCUUAAUCAAGA .(((((......(..(((((.............((((((.((((....)))).))))))..((((.((((((((......)))))))))))))))))..)....)))))........... (-33.30 = -33.80 + 0.50)

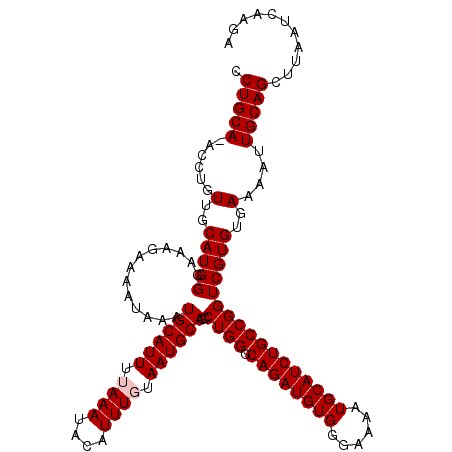
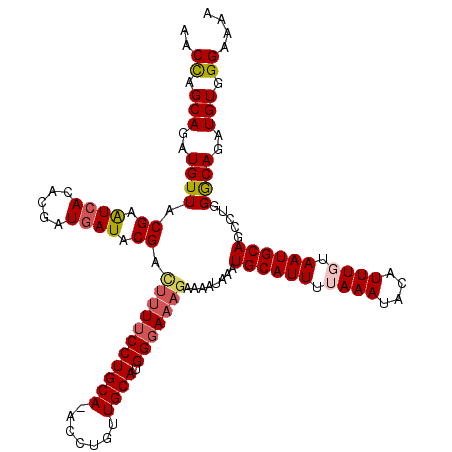
| Location | 9,085,570 – 9,085,689 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -23.64 |
| Energy contribution | -24.70 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9085570 119 - 20766785 AACCAGCAGAUGUUACGAAUCACACGAUGAUACGACUUUUCCUGCA-ACCUGUUGCAUGGGAAUAAAAAUAAAUGCAUUUUAAAUACAUUUGUAAUGCAGCCUGGACAGAUGUGGGAAAA ..((.(((..((((.((.((((.....)))).))...(((((((((-(....))))).)))))..........((((((.((((....)))).)))))).....))))..))).)).... ( -28.30) >DroSec_CAF1 50343 119 - 1 AACUAGCAGAUGUUACGAAUCACACGAUGAUACGGCUUUUCCUGCA-ACCUGUUGCAUGGGAAAGAAAAUAAAUGCAUUUAAAAUACAUUUGAAAUGCAGCCUGGGCAGAUGUGGGAAAA ..((.(((..((((..(.((((.....)))).)(((((((((((((-(....))))).)))))).........(((((((.(((....))).))))))))))..))))..))).)).... ( -30.30) >DroSim_CAF1 52556 119 - 1 AACCAGCAGAUGUUACGAAUCACACGAUGAUACGGCUUUUCCUGCA-ACCUGUUGCAUGGGAAAGAAAAUAAAUGCAUUUUAAAUACAUUUGUAAUGCAGCCUGGGCAGAUGUGGGAAAA ..((.(((..((((..(.((((.....)))).)(((((((((((((-(....))))).)))))).........((((((.((((....)))).)))))))))..))))..))).)).... ( -32.20) >DroEre_CAF1 57669 120 - 1 AACCAGCAGAUGUUACGAGUCACACGAUUAUACGAUUCUUCCUGCAAACCUGUUGCAUGGCAAAGAAAAUAAAUGCAUUUUAAAUAUUUUUCUAAUGCAGCCUGGGCAGAUGUGGGAAAA ................(((((............)))))((((..((...(((((.((.(((............((((((..(((....)))..)))))))))))))))).))..)))).. ( -24.90) >consensus AACCAGCAGAUGUUACGAAUCACACGAUGAUACGACUUUUCCUGCA_ACCUGUUGCAUGGGAAAGAAAAUAAAUGCAUUUUAAAUACAUUUGUAAUGCAGCCUGGGCAGAUGUGGGAAAA ..((.(((..((((.((.((((.....)))).)).(((((((((((.......)))).)))))))........((((((.((((....)))).)))))).....))))..))).)).... (-23.64 = -24.70 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:34 2006