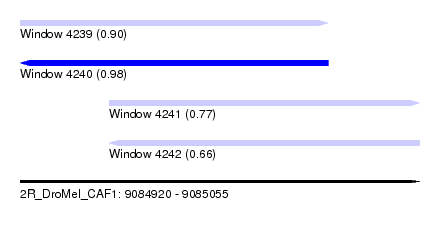
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,084,920 – 9,085,055 |
| Length | 135 |
| Max. P | 0.976189 |

| Location | 9,084,920 – 9,085,024 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9084920 104 + 20766785 AUUUUUCACACGAUCAGACUAU----------AGUAGGUUUACAAUCUGCAUAGGAAAAAAAUAAACGAUCGUCUUUUAUUUUCCCAUGCAACAGGUUUUAGGAUGAAAAUUAA ..((((((..((((((((((..----------....)))))......(((((.((((((.((...((....))...)).)))))).)))))...))))...)..)))))).... ( -18.70) >DroSec_CAF1 49691 102 + 1 AUUUUUCACACGAUCUGA-UAU----------AUAAGGUUUACAAUCUGCAUGGGAA-AAAAUAAACGACCGCCUUUUAUUUUCCCAUGCAACAGGUUUUGGAAUGAAAAUGAA ..((((((((.((((((.-...----------...((((.....))))(((((((((-(..(((((.(.....).)))))))))))))))..)))))).))...)))))).... ( -26.20) >DroSim_CAF1 51908 112 + 1 GUUUUUCACACGAUCAGA-UAUUAUAUAUCAGAUUAGGUUUACAAUCUGCAUGGGAA-AAUAUAAAGGACCGACUUUUAUUUUCCCAUGCAACAGGUUUUAGGAUGAAAAUGAA ..((((((..(((((.((-(((...))))).))))............((((((((((-((((.((((......))))))))))))))))))..........)..)))))).... ( -32.10) >consensus AUUUUUCACACGAUCAGA_UAU__________AUUAGGUUUACAAUCUGCAUGGGAA_AAAAUAAACGACCGACUUUUAUUUUCCCAUGCAACAGGUUUUAGGAUGAAAAUGAA ..((((((..(((((....................((((.....))))(((((((((...((((((.(.....).)))))))))))))))....))))...)..)))))).... (-18.00 = -18.67 + 0.67)

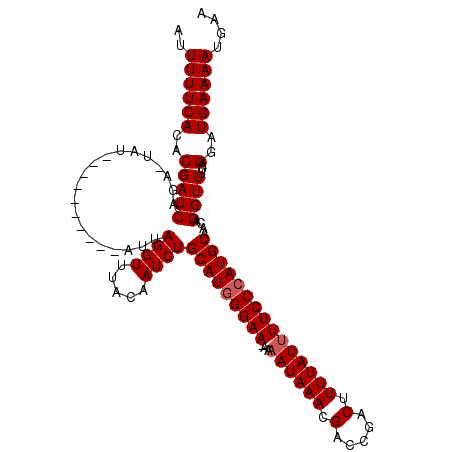
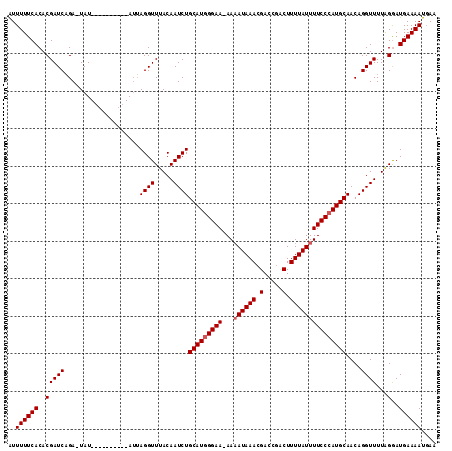
| Location | 9,084,920 – 9,085,024 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -20.51 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9084920 104 - 20766785 UUAAUUUUCAUCCUAAAACCUGUUGCAUGGGAAAAUAAAAGACGAUCGUUUAUUUUUUUCCUAUGCAGAUUGUAAACCUACU----------AUAGUCUGAUCGUGUGAAAAAU ....((((((..(...........(((((((((((.((.((((....)))).)).)))))))))))((((((((.......)----------)))))))....)..)))))).. ( -28.50) >DroSec_CAF1 49691 102 - 1 UUCAUUUUCAUUCCAAAACCUGUUGCAUGGGAAAAUAAAAGGCGGUCGUUUAUUUU-UUCCCAUGCAGAUUGUAAACCUUAU----------AUA-UCAGAUCGUGUGAAAAAU ....(((((((.(..........((((((((((((....((((....))))...))-))))))))))(((((((.....)))----------).)-)).....).))))))).. ( -25.10) >DroSim_CAF1 51908 112 - 1 UUCAUUUUCAUCCUAAAACCUGUUGCAUGGGAAAAUAAAAGUCGGUCCUUUAUAUU-UUCCCAUGCAGAUUGUAAACCUAAUCUGAUAUAUAAUA-UCUGAUCGUGUGAAAAAC ....((((((..(....((...(((((((((((((((((((......)))).))))-)))))))))))...)).......(((.(((((...)))-)).))).)..)))))).. ( -31.10) >consensus UUCAUUUUCAUCCUAAAACCUGUUGCAUGGGAAAAUAAAAGACGGUCGUUUAUUUU_UUCCCAUGCAGAUUGUAAACCUAAU__________AUA_UCUGAUCGUGUGAAAAAU ....(((((((.(..........((((((((((((((((.(.....).))))))...))))))))))((((............................))))).))))))).. (-20.51 = -20.62 + 0.11)

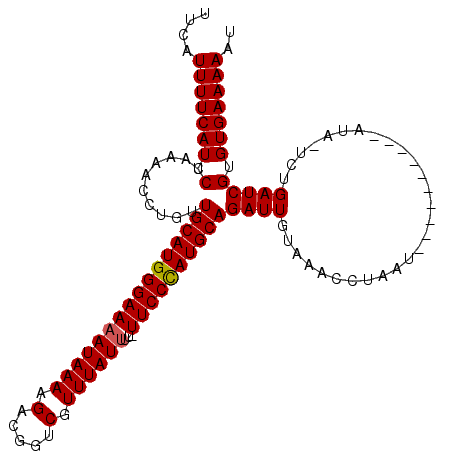
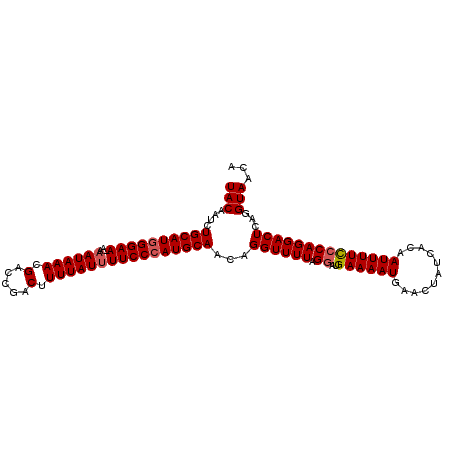
| Location | 9,084,950 – 9,085,055 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9084950 105 + 20766785 UACAAUCUGCAUAGGAAAAAAAUAAACGAUCGUCUUUUAUUUUCCCAUGCAACAGGUUUUAGGAUGAAAAUUAAAUAUCACAAUUUUUCCAGGACUCAGGUAACA (((....(((((.((((((.((...((....))...)).)))))).)))))...((((((.(((..((((((.........)))))))))))))))...)))... ( -17.80) >DroSec_CAF1 49720 104 + 1 UACAAUCUGCAUGGGAA-AAAAUAAACGACCGCCUUUUAUUUUCCCAUGCAACAGGUUUUGGAAUGAAAAUGAACUAUCAUAAUUUUCCCAGGACUCAGGUAACA (((....((((((((((-(..(((((.(.....).))))))))))))))))...((((((((...((((((...........))))))))))))))...)))... ( -27.90) >DroSim_CAF1 51947 104 + 1 UACAAUCUGCAUGGGAA-AAUAUAAAGGACCGACUUUUAUUUUCCCAUGCAACAGGUUUUAGGAUGAAAAUGAACUAUCACAAUUUUCCCAGGACUCAGGUAACA (((....((((((((((-((((.((((......))))))))))))))))))...((((((.(((......(((....)))......))).))))))...)))... ( -31.20) >consensus UACAAUCUGCAUGGGAA_AAAAUAAACGACCGACUUUUAUUUUCCCAUGCAACAGGUUUUAGGAUGAAAAUGAACUAUCACAAUUUUCCCAGGACUCAGGUAACA (((....((((((((((...((((((.(.....).))))))))))))))))...((((((.((..((((((...........))))))))))))))...)))... (-19.12 = -19.90 + 0.78)


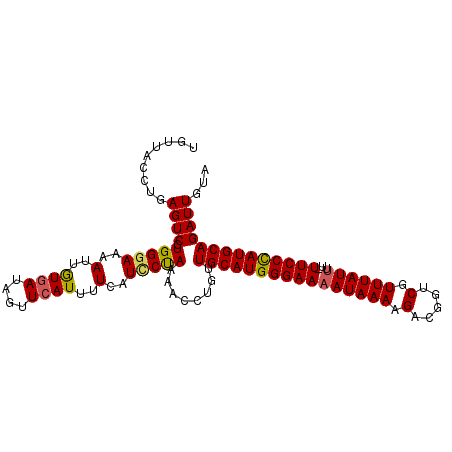
| Location | 9,084,950 – 9,085,055 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9084950 105 - 20766785 UGUUACCUGAGUCCUGGAAAAAUUGUGAUAUUUAAUUUUCAUCCUAAAACCUGUUGCAUGGGAAAAUAAAAGACGAUCGUUUAUUUUUUUCCUAUGCAGAUUGUA .........((((..((((((((((.......)))))))..)))..........((((((((((((.((.((((....)))).)).))))))))))))))))... ( -24.30) >DroSec_CAF1 49720 104 - 1 UGUUACCUGAGUCCUGGGAAAAUUAUGAUAGUUCAUUUUCAUUCCAAAACCUGUUGCAUGGGAAAAUAAAAGGCGGUCGUUUAUUUU-UUCCCAUGCAGAUUGUA .........((((.(((((..(..((((....))))..)..)))))........((((((((((((....((((....))))...))-))))))))))))))... ( -27.60) >DroSim_CAF1 51947 104 - 1 UGUUACCUGAGUCCUGGGAAAAUUGUGAUAGUUCAUUUUCAUCCUAAAACCUGUUGCAUGGGAAAAUAAAAGUCGGUCCUUUAUAUU-UUCCCAUGCAGAUUGUA .........((((.(((((..(..((((....))))..)..)))))........((((((((((((((((((......)))).))))-))))))))))))))... ( -30.90) >consensus UGUUACCUGAGUCCUGGGAAAAUUGUGAUAGUUCAUUUUCAUCCUAAAACCUGUUGCAUGGGAAAAUAAAAGACGGUCGUUUAUUUU_UUCCCAUGCAGAUUGUA .........((((.(((((..(..((((....))))..)..)))))........((((((((((((((((.(.....).))))))...))))))))))))))... (-20.90 = -21.13 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:29 2006