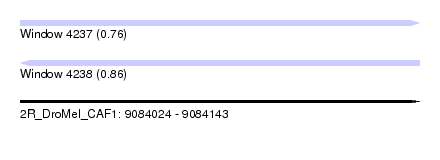
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,084,024 – 9,084,143 |
| Length | 119 |
| Max. P | 0.858224 |

| Location | 9,084,024 – 9,084,143 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -14.02 |
| Energy contribution | -13.86 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
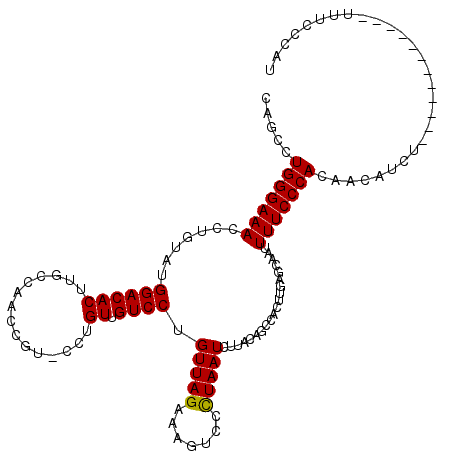
>2R_DroMel_CAF1 9084024 119 + 20766785 CAGCCUGGGAAACCUUUAUGGACACUUAUCAACUGU-CCUGUAGUCCUGUUAGAAAAUCCCUAAUCUUACAGUCACUUGAGCAAUUUUCCCACAACAUAUUUUGGGAGCAAUUUUCCCAC .....(((((((((.....)).......((((.((.-.((((((....(((((.......))))).)))))).)).)))).....)))))))..........((((((.....)))))). ( -26.00) >DroSec_CAF1 48803 107 + 1 CAGCCUGGGAAACAUGUAUGGACACUUGCCCACUGU-CCUGUUGUCCUGUUAGAAAGUCCCUAAUCUUACAGCCACUUGAGCAAUUUUCCCACAACAUCU------------UUUCCCAU .....(((((((.((((..(((.(.((((.((..((-...(((((...(((((.......)))))...))))).)).)).)))).).)))....))))..------------))))))). ( -23.60) >DroSim_CAF1 51041 108 + 1 CAGCCUGGGAAAUCUGUAUGGACACUUGCCCACAGAACUUGUUGUCCUGUUAGAAAGUCCUUAAUCUUACAGCCACUUGAGCAAUUUUCCCACAACUUCU------------UUUCCCAU .....((((((((((((..((.(....))).)))))....(((((..(((.(((..........))).)))(((....).)).........)))))....------------))))))). ( -20.40) >DroEre_CAF1 56242 107 + 1 CGACUUGGGAAAGGCCUAUGGACACUUGCCAGCCGU-CCUGUUGUCCUGUUAGUUAGUCCCUAAUCCAACAGCCACUUGAGGCCUUUUCCCACGACAACU------------UUGCCCAU ...(.((((((((((((..((((...........))-)).(((((...(((((.......)))))...)))))......))))).))))))).)......------------........ ( -29.40) >DroYak_CAF1 54085 107 + 1 UCACUUGGGAAACCUUUACGGACACUUGUCAACCGU-CCUGUUGUCCUGUUAGAUAGUUACUAAUCUCAUAGUCACUUCAGGCCGUUUCCCACGGCAACU------------UUUCCCAU .....(((((((.......((((((..(.(....).-)..).))))).(((.((.(((.((((......)))).)))))..(((((.....)))))))).------------))))))). ( -27.00) >consensus CAGCCUGGGAAACCUGUAUGGACACUUGCCAACCGU_CCUGUUGUCCUGUUAGAAAGUCCCUAAUCUUACAGCCACUUGAGCAAUUUUCCCACAACAUCU____________UUUCCCAU .....(((((((.......((((((...............)).)))).(((((.......)))))....................)))))))............................ (-14.02 = -13.86 + -0.16)

| Location | 9,084,024 – 9,084,143 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.54 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
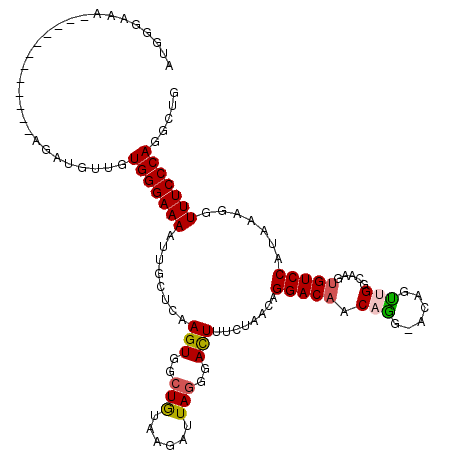
>2R_DroMel_CAF1 9084024 119 - 20766785 GUGGGAAAAUUGCUCCCAAAAUAUGUUGUGGGAAAAUUGCUCAAGUGACUGUAAGAUUAGGGAUUUUCUAACAGGACUACAGG-ACAGUUGAUAAGUGUCCAUAAAGGUUUCCCAGGCUG .(((((.......)))))......(((.(((((((........(((..((((.(((..(....)..))).)))).)))...((-(((.(.....).))))).......)))))))))).. ( -30.10) >DroSec_CAF1 48803 107 - 1 AUGGGAAA------------AGAUGUUGUGGGAAAAUUGCUCAAGUGGCUGUAAGAUUAGGGACUUUCUAACAGGACAACAGG-ACAGUGGGCAAGUGUCCAUACAUGUUUCCCAGGCUG .(((((((------------..((((.(((((....(((((((..((.((((.(((..........))).))))..)).....-....)))))))...))))))))).)))))))..... ( -32.60) >DroSim_CAF1 51041 108 - 1 AUGGGAAA------------AGAAGUUGUGGGAAAAUUGCUCAAGUGGCUGUAAGAUUAAGGACUUUCUAACAGGACAACAAGUUCUGUGGGCAAGUGUCCAUACAGAUUUCCCAGGCUG .(((((((------------.....((((((((...((((((...........(((..........))).(((((((.....)))))))))))))...))).))))).)))))))..... ( -32.40) >DroEre_CAF1 56242 107 - 1 AUGGGCAA------------AGUUGUCGUGGGAAAAGGCCUCAAGUGGCUGUUGGAUUAGGGACUAACUAACAGGACAACAGG-ACGGCUGGCAAGUGUCCAUAGGCCUUUCCCAAGUCG ...((((.------------...)))).(((((((.(((((....((.(((((((.((((...)))))))))))..))...((-(((.((....)))))))..))))))))))))..... ( -38.50) >DroYak_CAF1 54085 107 - 1 AUGGGAAA------------AGUUGCCGUGGGAAACGGCCUGAAGUGACUAUGAGAUUAGUAACUAUCUAACAGGACAACAGG-ACGGUUGACAAGUGUCCGUAAAGGUUUCCCAAGUGA .(((((((------------.((((((((.(....).))..(((((.((((......)))).))).)).....)).)))).((-(((.(.....).))))).......)))))))..... ( -32.00) >consensus AUGGGAAA____________AGAUGUUGUGGGAAAAUUGCUCAAGUGGCUGUAAGAUUAGGGACUUUCUAACAGGACAACAGG_ACAGUUGGCAAGUGUCCAUAAAGGUUUCCCAGGCUG ............................(((((((........(((..(((......)))..)))........(((((.(((......))).....))))).......)))))))..... (-15.38 = -15.54 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:26 2006