
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,083,165 – 9,083,285 |
| Length | 120 |
| Max. P | 0.999386 |

| Location | 9,083,165 – 9,083,285 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.66 |
| Mean single sequence MFE | -34.94 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9083165 120 + 20766785 UUAGUAUUCCAUAAAUAGCCCGCAUUCUCACGCCUUCGUGGAAUCGGAGAACAUGCGGGCCAAACGAACCCAAAGGAAUCCAUAAAAAGAUCAAAGAUAUCCUAAGGUUUCGCGUAUCAU .................((((((((((((...((.....)).....))))..))))))))....((((.((..(((((((...............))).))))..)).))))........ ( -30.96) >DroSec_CAF1 47961 120 + 1 UUAGUAUUCCAUAUAUAGCCCGCAUUCUCACGCCUUCGGGGAAUCGGAGAACAUGCGGGCCAAACGAACCCAAAGGAAUCCAUAAAAAGAUCAAAGAUAUCCUAAGGUUUCGCGUAUCAU .................((((((((((((.((((....))....))))))..))))))))....((((.((..(((((((...............))).))))..)).))))........ ( -33.16) >DroSim_CAF1 50217 120 + 1 UUGGUAUUCCAUAUGUAGCCCGCAUUCUCACGCCUUCGGGGAAUCGGAGAACAUGCGGGCCAAACGAACCCAAAGGAAUCCAUAAAAAGAUCAAAGAUAUCCUAAGGUUUCGCGUAUCAU .((((((..........((((((((((((.((((....))....))))))..))))))))....((((.((..(((((((...............))).))))..)).)))).)))))). ( -36.46) >DroEre_CAF1 55391 119 + 1 UUAGUAUUCCAUAUGUAGCCCGCAUUCUCACGCCUUCGGGCAAUCG-AGAACAUGCGGGCCAAACGAACCCAAAGGAAUCCAUAAAAAGAUCAAAGAUAUCCUAAGGUUUCGCGUAUCAU ..........((((((.((((((((((((..(((....)))....)-)))..)))))))).....(((.((..(((((((...............))).))))..)).)))))))))... ( -37.46) >DroYak_CAF1 53272 119 + 1 UUAGUAUUCUACAUGUAGCCCGCAUUCUCACGCCCUCGGGGCAUCG-GGAACAUGCGGGCCAAGUGAACCGAAAGGAAUCCAUAAAAAGAUCAAAGAUAUCCUAAGGUUUCGCGUAUCGU .........(((.....((((((((((((..((((...))))...)-)))..))))))))...(((((((...(((((((...............))).))))..)).)))))))).... ( -36.66) >consensus UUAGUAUUCCAUAUGUAGCCCGCAUUCUCACGCCUUCGGGGAAUCGGAGAACAUGCGGGCCAAACGAACCCAAAGGAAUCCAUAAAAAGAUCAAAGAUAUCCUAAGGUUUCGCGUAUCAU .................((((((((((((...((.....)).....))))..))))))))....((((.((..(((((((...............))).))))..)).))))........ (-26.22 = -26.70 + 0.48)

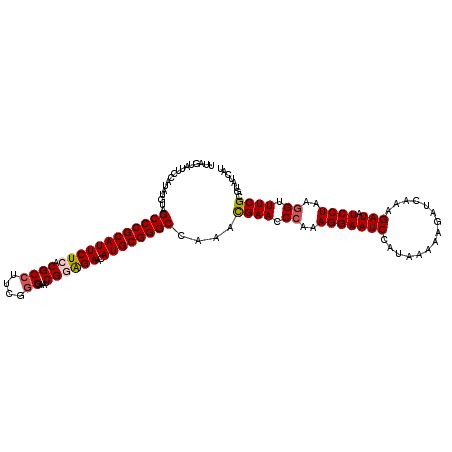

| Location | 9,083,165 – 9,083,285 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.66 |
| Mean single sequence MFE | -41.22 |
| Consensus MFE | -35.26 |
| Energy contribution | -34.86 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.999386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9083165 120 - 20766785 AUGAUACGCGAAACCUUAGGAUAUCUUUGAUCUUUUUAUGGAUUCCUUUGGGUUCGUUUGGCCCGCAUGUUCUCCGAUUCCACGAAGGCGUGAGAAUGCGGGCUAUUUAUGGAAUACUAA .......(((((.((..((((.((((.(((.....))).))))))))..)).))))).((((((((((..((((((.(((...)))..)).))))))))))))))............... ( -40.40) >DroSec_CAF1 47961 120 - 1 AUGAUACGCGAAACCUUAGGAUAUCUUUGAUCUUUUUAUGGAUUCCUUUGGGUUCGUUUGGCCCGCAUGUUCUCCGAUUCCCCGAAGGCGUGAGAAUGCGGGCUAUAUAUGGAAUACUAA .......(((((.((..((((.((((.(((.....))).))))))))..)).))))).((((((((((..((((((.(((...)))..)).))))))))))))))............... ( -40.40) >DroSim_CAF1 50217 120 - 1 AUGAUACGCGAAACCUUAGGAUAUCUUUGAUCUUUUUAUGGAUUCCUUUGGGUUCGUUUGGCCCGCAUGUUCUCCGAUUCCCCGAAGGCGUGAGAAUGCGGGCUACAUAUGGAAUACCAA (((....(((((.((..((((.((((.(((.....))).))))))))..)).))))).((((((((((..((((((.(((...)))..)).))))))))))))))))).(((....))). ( -42.20) >DroEre_CAF1 55391 119 - 1 AUGAUACGCGAAACCUUAGGAUAUCUUUGAUCUUUUUAUGGAUUCCUUUGGGUUCGUUUGGCCCGCAUGUUCU-CGAUUGCCCGAAGGCGUGAGAAUGCGGGCUACAUAUGGAAUACUAA (((....(((((.((..((((.((((.(((.....))).))))))))..)).))))).((((((((((..(((-((...(((....))).))))))))))))))))))............ ( -43.60) >DroYak_CAF1 53272 119 - 1 ACGAUACGCGAAACCUUAGGAUAUCUUUGAUCUUUUUAUGGAUUCCUUUCGGUUCACUUGGCCCGCAUGUUCC-CGAUGCCCCGAGGGCGUGAGAAUGCGGGCUACAUGUAGAAUACUAA ....((((.(((.((..((((.((((.(((.....))).))))))))...)))))...((((((((((.(((.-..((((((...))))))))).))))))))))..))))......... ( -39.50) >consensus AUGAUACGCGAAACCUUAGGAUAUCUUUGAUCUUUUUAUGGAUUCCUUUGGGUUCGUUUGGCCCGCAUGUUCUCCGAUUCCCCGAAGGCGUGAGAAUGCGGGCUACAUAUGGAAUACUAA .......(((((.((..((((.((((.(((.....))).))))))))..)).))))).((((((((((..(((.((......)).)))(....).))))))))))....(((....))). (-35.26 = -34.86 + -0.40)

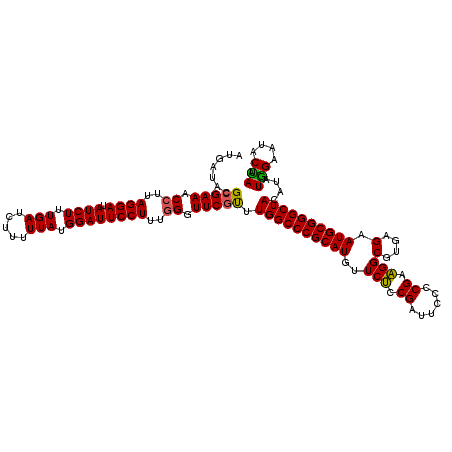
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:23 2006