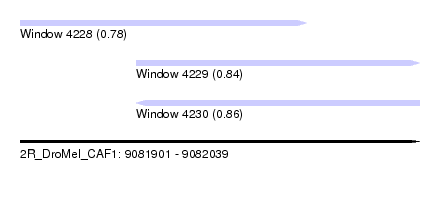
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,081,901 – 9,082,039 |
| Length | 138 |
| Max. P | 0.856204 |

| Location | 9,081,901 – 9,082,000 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.74 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -18.35 |
| Energy contribution | -19.72 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9081901 99 + 20766785 GCCAAAGCGAAAAAAAGGGACAGGGAAACAAUUAACAACUAAUUUAGGCCUCUGCAUAUAAUUUUAUGGAGAGUGGUGCGGAGAGUCGCAGUCCCACAG------- ((....))........(((((..(....).................(((((((((((...(((((....))))).)))))))).)))...)))))....------- ( -29.70) >DroSec_CAF1 46658 95 + 1 GCCAAAGCGUAA-AAAGGGACAGGGAAACAAUUAACAACUAAUUUAGGCCUCGGCAUAUAAUUUUAUGGAGAG---UGCGGAGAGUCGCAGUCCCACAG------- ((....))....-...(((((..(....).................((((((.((((...............)---))).))).)))...)))))....------- ( -24.16) >DroSim_CAF1 48928 95 + 1 GCCAAAGCGAAA-AAAGGGACAGGGAAACAAUUAACAACUAAUUUAGGCCUCUGCAUAUAAUUUUAUGGAGAG---UGCGGAGAGUCGCCGUCCCACAG------- ((....))....-...(((((..(....).................(((((((((((...............)---))))))).)))...)))))....------- ( -26.36) >DroEre_CAF1 54078 95 + 1 GCCAAAGCGAA--AAAGGGCCAGGGAAACAAUUAACAACUAAUUUAGGCCUAUGCAUAUCAUUUUACGGAGAG---UGAAGAGAGU------CCCACGAGGAGAUG ......(((..--...(((((..(....)(((((.....)))))..))))).)))...((.((((((.....)---))))).))((------(((....)).))). ( -18.70) >consensus GCCAAAGCGAAA_AAAGGGACAGGGAAACAAUUAACAACUAAUUUAGGCCUCUGCAUAUAAUUUUAUGGAGAG___UGCGGAGAGUCGCAGUCCCACAG_______ ((....))........(((((..(....).................((((((((((.....((((...))))....))))))).)))...)))))........... (-18.35 = -19.72 + 1.37)

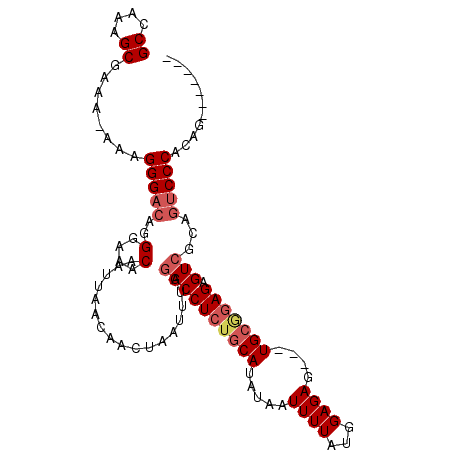
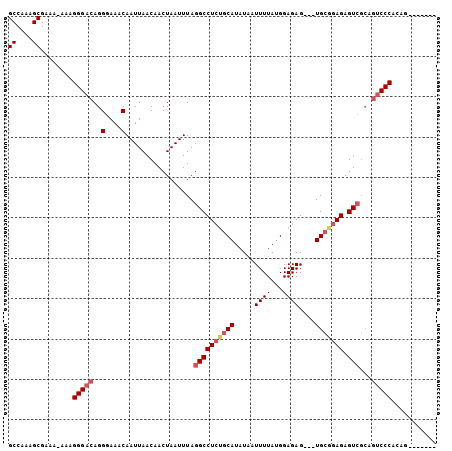
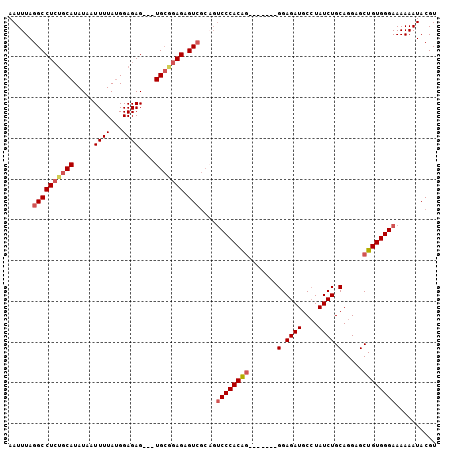
| Location | 9,081,941 – 9,082,039 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -21.86 |
| Energy contribution | -23.05 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9081941 98 + 20766785 AAUUUAGGCCUCUGCAUAUAAUUUUAUGGAGAGUGGUGCGGAGAGUCGCAGUCCCACAG-------GGAGAUGCCUAUCUGCAGGAGCUGUGGGAAAAAAUACGU ......(((((((((((...(((((....))))).)))))))).)))....((((((((-------.....(((......)))....)))))))).......... ( -33.70) >DroSec_CAF1 46697 95 + 1 AAUUUAGGCCUCGGCAUAUAAUUUUAUGGAGAG---UGCGGAGAGUCGCAGUCCCACAG-------GGAGAUGCCUAUCUGCAGGAGCUGUGGGAAAAAAUACGU ......((((((.((((...............)---))).))).)))....((((((((-------.....(((......)))....)))))))).......... ( -28.16) >DroSim_CAF1 48967 95 + 1 AAUUUAGGCCUCUGCAUAUAAUUUUAUGGAGAG---UGCGGAGAGUCGCCGUCCCACAG-------GGAGAUGCAUAUCUGCAGGAGCUGUGGGAAAAAAUACGU ......(((((((((((...............)---))))))).)))....((((((((-------.....((((....))))....)))))))).......... ( -31.66) >DroEre_CAF1 54116 96 + 1 AAUUUAGGCCUAUGCAUAUCAUUUUACGGAGAG---UGAAGAGAGU------CCCACGAGGAGAUGGGAGAUGCCUAUCUGCAGGAGCUGUGGGAAAAAAUACGU .......((....))...((.((((((.....)---))))).)).(------((((((...(((((((.....)))))))((....))))))))).......... ( -24.60) >consensus AAUUUAGGCCUCUGCAUAUAAUUUUAUGGAGAG___UGCGGAGAGUCGCAGUCCCACAG_______GGAGAUGCCUAUCUGCAGGAGCUGUGGGAAAAAAUACGU ......((((((((((.....((((...))))....))))))).)))....((((((((.......(.((((....)))).).....)))))))).......... (-21.86 = -23.05 + 1.19)

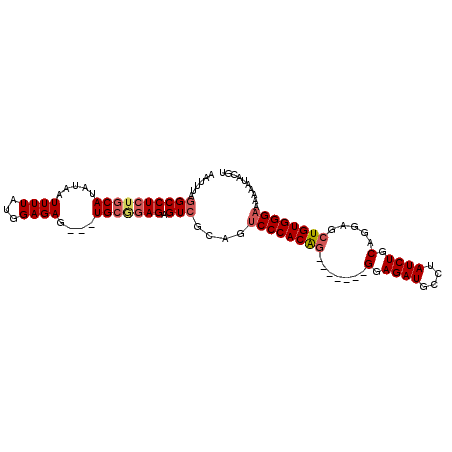
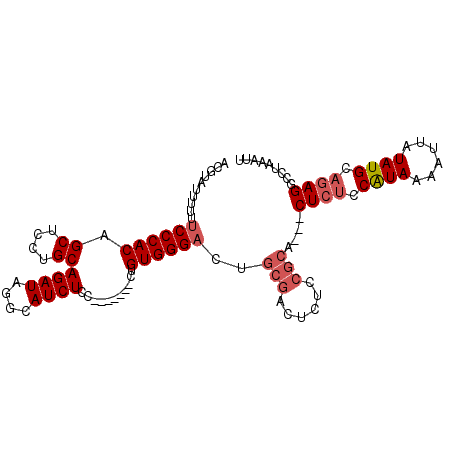
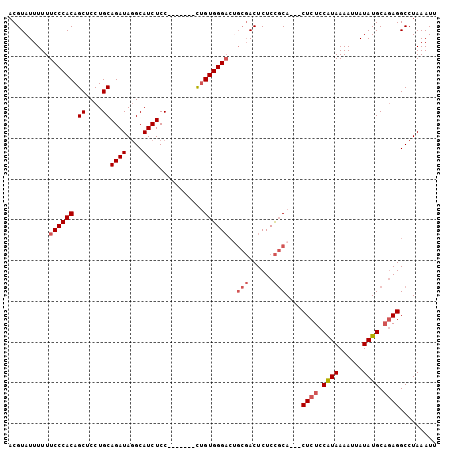
| Location | 9,081,941 – 9,082,039 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -16.29 |
| Energy contribution | -17.60 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9081941 98 - 20766785 ACGUAUUUUUUCCCACAGCUCCUGCAGAUAGGCAUCUCC-------CUGUGGGACUGCGACUCUCCGCACCACUCUCCAUAAAAUUAUAUGCAGAGGCCUAAAUU ..........((((((((....(((......))).....-------)))))))).((((......))))...((((.((((......)))).))))......... ( -26.40) >DroSec_CAF1 46697 95 - 1 ACGUAUUUUUUCCCACAGCUCCUGCAGAUAGGCAUCUCC-------CUGUGGGACUGCGACUCUCCGCA---CUCUCCAUAAAAUUAUAUGCCGAGGCCUAAAUU ..........((((((((....(((......))).....-------)))))))).((((......))))---(((..((((......))))..)))......... ( -24.20) >DroSim_CAF1 48967 95 - 1 ACGUAUUUUUUCCCACAGCUCCUGCAGAUAUGCAUCUCC-------CUGUGGGACGGCGACUCUCCGCA---CUCUCCAUAAAAUUAUAUGCAGAGGCCUAAAUU .(((......((((((((....((((....)))).....-------))))))))..))).......((.---((((.((((......)))).))))))....... ( -27.80) >DroEre_CAF1 54116 96 - 1 ACGUAUUUUUUCCCACAGCUCCUGCAGAUAGGCAUCUCCCAUCUCCUCGUGGG------ACUCUCUUCA---CUCUCCGUAAAAUGAUAUGCAUAGGCCUAAAUU .................((....))...(((((...((((((......)))))------).........---......(((........)))....))))).... ( -17.00) >consensus ACGUAUUUUUUCCCACAGCUCCUGCAGAUAGGCAUCUCC_______CUGUGGGACUGCGACUCUCCGCA___CUCUCCAUAAAAUUAUAUGCAGAGGCCUAAAUU ..........((((((.((....))((((....))))...........))))))..(((......)))....((((.((((......)))).))))......... (-16.29 = -17.60 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:18 2006