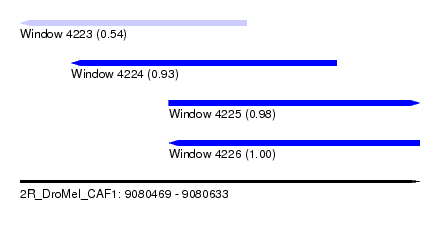
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,080,469 – 9,080,633 |
| Length | 164 |
| Max. P | 0.997002 |

| Location | 9,080,469 – 9,080,562 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.11 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9080469 93 - 20766785 CAUCAGCCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCAAAAAUGGCCCGCGGCAAAGGAAACGCCCCUCGGCCGCACUCU-ACU------------------CUUCAUAU ....................((((........(((((........)))))(((((..(((.......)))..)))))..)))-)..------------------........ ( -25.20) >DroSec_CAF1 45273 86 - 1 CAUCAGCCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA--AAUGGCCCGCGGCAAAGGAGACGCCCCUCGGCCGCAC------U------------------CUCCACAU ....................((((........(((((...--...)))))(((((..(((.......)))..)))))..------.------------------.))))... ( -27.70) >DroSim_CAF1 47552 78 - 1 CAUCAGCCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA--AAUGGCCCGCGGCAAAGGAAACGCCCCC--------C------U------------------CUCCAUAU ....................((((........(((((...--...)))))..(((...(....))))...--------.------.------------------.))))... ( -17.30) >DroEre_CAF1 52609 105 - 1 CAUCAGGCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA--AAUGGCCCGCGGCAAAGGAAACGCCCCUC---CGAACUCUCUCUCUCU--CACACUCGCUAACUCCAUAU .....(((.((..........(((........(((((...--...)))))..(((...(....))))..))---)((......)).....--....)).))).......... ( -19.70) >DroYak_CAF1 50420 106 - 1 CAUCAGGCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA--AAUGGCCCGCGGCCAAGGAAACGCCCCUC---CGCACUCUCACUCCCUUUGACAUACAUGGACG-GAUAU .(((...(.......((((..(((........(((((...--...)))))((((....(....)......)---)))........)))..))))........)...-))).. ( -23.86) >consensus CAUCAGCCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA__AAUGGCCCGCGGCAAAGGAAACGCCCCUC___CGCACUCU__CU__________________CUCCAUAU .....................((.........(((((........)))))..(((...(....))))))........................................... (-14.52 = -14.52 + -0.00)

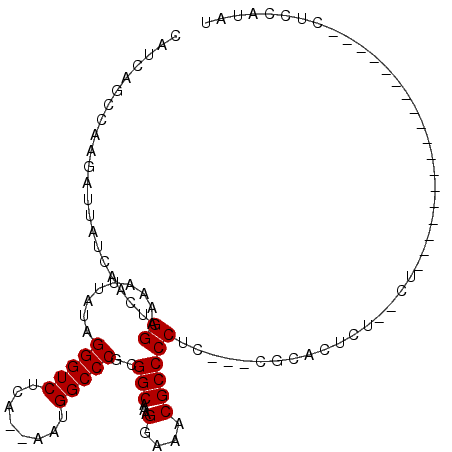
| Location | 9,080,490 – 9,080,599 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 96.65 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.22 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9080490 109 - 20766785 CCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGCCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCAAAAAUGGCCCGCGGCAAAGGAAACGCCCCUCG ....(((((((((((.....))).(((.......(((.(((......))).)))...............(((((........)))))..)))..))).)))))...... ( -29.40) >DroSec_CAF1 45289 107 - 1 CCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGCCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA--AAUGGCCCGCGGCAAAGGAGACGCCCCUCG ....(((((((((((.....))).(((.......(((.(((......))).)))...............(((((...--...)))))..)))..)))).))))...... ( -29.60) >DroSim_CAF1 47562 105 - 1 CCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGCCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA--AAUGGCCCGCGGCAAAGGAAACGCCCCC-- ....(((((((((((.....))).(((.......(((.(((......))).)))...............(((((...--...)))))..)))..))).)))))....-- ( -29.60) >DroEre_CAF1 52645 106 - 1 CCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGGCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA--AAUGGCCCGCGGCAAAGGAAACGCCCCUC- ....(((((((((((.....))).(((.......(((.(((......))).)))...............(((((...--...)))))..)))..))).))))).....- ( -29.60) >DroYak_CAF1 50457 106 - 1 CCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGGCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA--AAUGGCCCGCGGCCAAGGAAACGCCCCUC- ....(((((((((((.....))).(((.......(((.(((......))).)))...............(((((...--...)))))..)))..))).))))).....- ( -28.70) >consensus CCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGCCAAGAUUAUCAACUGGAAAAUUAUAGGGUCUCA__AAUGGCCCGCGGCAAAGGAAACGCCCCUC_ ....(((((((((((.....))).(((.......(((.(((......))).)))...............(((((........)))))..)))..))))).)))...... (-29.38 = -29.22 + -0.16)


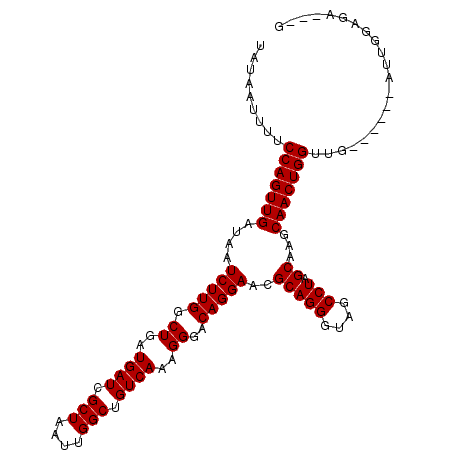
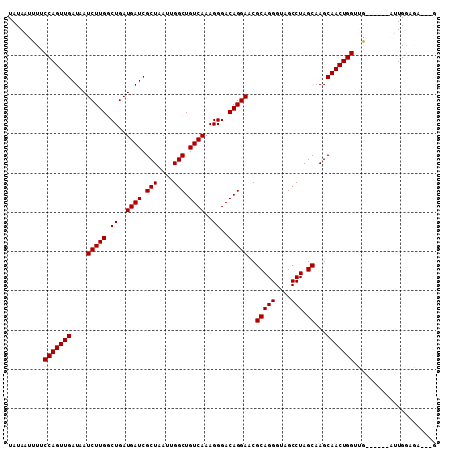
| Location | 9,080,530 – 9,080,633 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -26.32 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9080530 103 + 20766785 UAUAAUUUUCCAGUUGAUAAUCUUGGCUGAUGAUCGCUAAUUGGCUGUCAAAGGGACAGGAACGCAGGGUAGCCUAGCAAGCAACUGGUUGAUG---AUUGGAGA---G ....(((..(((((((....(((((.((..((((.(((....))).))))..))..)))))..(((((....))).))...)))))))..))).---........---. ( -29.50) >DroSec_CAF1 45327 100 + 1 UAUAAUUUUCCAGUUGAUAAUCUUGGCUGAUGAUCGCUAAUUGGCUGUCAAAGGGACAGGAACGCAGGGUAGCCUAGCAAGCAACUGGUUG------AUUGCAGA---G ..(((((..(((((((....(((((.((..((((.(((....))).))))..))..)))))..(((((....))).))...)))))))..)------))))....---. ( -31.20) >DroSim_CAF1 47598 100 + 1 UAUAAUUUUCCAGUUGAUAAUCUUGGCUGAUGAUCGCUAAUUGGCUGUCAAAGGGACAGGAACGCAGGGUAGCCUAGCAAGCAACUGGUUG------AUUGGAGA---G ..(((((..(((((((....(((((.((..((((.(((....))).))))..))..)))))..(((((....))).))...)))))))..)------))))....---. ( -31.50) >DroEre_CAF1 52682 101 + 1 UAUAAUUUUCCAGUUGAUAAUCUUGCCUGAUGAUCGCUAAUUGGCUGUCAAAGGGACAGGAACGCAGGGUAGCCUAGCAAGCAACUGGUGA--------UGGGCAACAA ....(((..(((((((....((((((((..((((.(((....))).))))..))).)))))..(((((....))).))...))))))).))--------).(....).. ( -32.50) >DroYak_CAF1 50494 105 + 1 UAUAAUUUUCCAGUUGAUAAUCUUGCCUGAUGAUCGCUAAUUGGCUGUCAAAGGGACAGGAACGCAGGGUCGCCUAGCAAGCAACUGGUGGAUCGAACUUGGGCA---- ....(((..(((((((....((((((((..((((.(((....))).))))..))).)))))..(((((....))).))...)))))))..)))............---- ( -32.80) >consensus UAUAAUUUUCCAGUUGAUAAUCUUGGCUGAUGAUCGCUAAUUGGCUGUCAAAGGGACAGGAACGCAGGGUAGCCUAGCAAGCAACUGGUUG______AUUGGAGA___G .........(((((((....(((((.((..((((.(((....))).))))..))..)))))..(((((....))).))...)))))))..................... (-26.32 = -26.32 + 0.00)

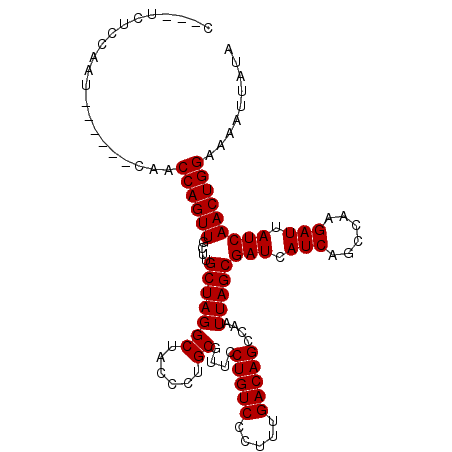
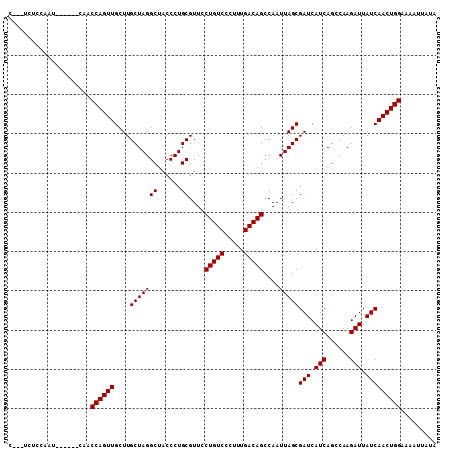
| Location | 9,080,530 – 9,080,633 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9080530 103 - 20766785 C---UCUCCAAU---CAUCAACCAGUUGCUUGCUAGGCUACCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGCCAAGAUUAUCAACUGGAAAAUUAUA .---........---......(((((((....((.((((......((((.(((((.....)))))......))))......)))).)).....)))))))......... ( -24.10) >DroSec_CAF1 45327 100 - 1 C---UCUGCAAU------CAACCAGUUGCUUGCUAGGCUACCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGCCAAGAUUAUCAACUGGAAAAUUAUA .---........------...(((((((....((.((((......((((.(((((.....)))))......))))......)))).)).....)))))))......... ( -24.10) >DroSim_CAF1 47598 100 - 1 C---UCUCCAAU------CAACCAGUUGCUUGCUAGGCUACCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGCCAAGAUUAUCAACUGGAAAAUUAUA .---........------...(((((((....((.((((......((((.(((((.....)))))......))))......)))).)).....)))))))......... ( -24.10) >DroEre_CAF1 52682 101 - 1 UUGUUGCCCA--------UCACCAGUUGCUUGCUAGGCUACCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGGCAAGAUUAUCAACUGGAAAAUUAUA ..........--------...((((((((((((((((....)))((....(((((.....))))).......))........)))))).....)))))))......... ( -23.80) >DroYak_CAF1 50494 105 - 1 ----UGCCCAAGUUCGAUCCACCAGUUGCUUGCUAGGCGACCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGGCAAGAUUAUCAACUGGAAAAUUAUA ----.................(((((((((((((.(..((.((((.....(((((.....))))).....))).).))..).)))))).....)))))))......... ( -25.10) >consensus C___UCUCCAAU______CAACCAGUUGCUUGCUAGGCUACCCUGCGUUCCUGUCCCUUUGACAGCCAAUUAGCGAUCAUCAGCCAAGAUUAUCAACUGGAAAAUUAUA .....................((((((....(((((((......))....(((((.....)))))....)))))(((.(((......))).)))))))))......... (-22.82 = -22.82 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:14 2006