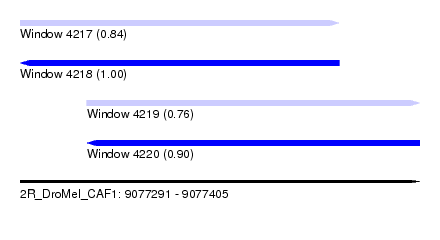
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,077,291 – 9,077,405 |
| Length | 114 |
| Max. P | 0.998241 |

| Location | 9,077,291 – 9,077,382 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.65 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9077291 91 + 20766785 GAUGCA-------CAUCCAUUGCAU--------------CCAGUGCAACGAAGUGCAACGCCAUCAUUUUUUCCCACAGCCGAGGGCUCUGGGUUAU--------AUUUAACAAAUGCAU ..((((-------(.((..((((((--------------...)))))).)).)))))..((............(((.((((...)))).)))((((.--------...))))....)).. ( -24.00) >DroSec_CAF1 42207 89 + 1 GUU--A-------CAUUCGUUGCAU--------------CCAGUGCAACCAAGUGCAACGCCAUGGAUCUUUCCCACAGCCGAGGGCUCUGGGUUAU--------AUUUAACAAAUGCAU (((--.-------((((.(((((((--------------...)))))))..)))).)))....(((((.(..((((.((((...)))).))))..).--------))))).......... ( -23.30) >DroSim_CAF1 44014 80 + 1 G------------------UUGCAU--------------CCAGUGCAACCAAGUGCAACGCCAUGGAUUUUUCCCACAGCCGAGGGCUCUGGGUUAU--------AUUUAACAAAUGCAU (------------------((((((--------------...)))))))...(((((......(((((....((((.((((...)))).))))....--------))))).....))))) ( -23.40) >DroEre_CAF1 49575 98 + 1 AAUGCAUCCAAUGCAUCCGAUGCAU--------------CCAGCGCAGCGAAGUGCAACGCCAUCAGUUUUUCCCACAGCCGAGGGCUCUGGGUUAU--------AUUUAACAAAUGCAU .((((((...))))))...((((((--------------(((((((......))))...(((.((.(((........))).)).)))..)))((((.--------...))))..)))))) ( -24.70) >DroYak_CAF1 45356 120 + 1 AAUGCAUCCAAUGCAUCCAAUGCAUCCACUGCAUCCCAUCCAGUGCAGCGAAGUGCAACGCCAUCAGUUUUUCCCACAGCCGAGGGCUCUGGGUUAUAUAUGUAUAUUUAUCAAAUGCAU .((((((...((((((....(((((.(.((((((........)))))).)..)))))...............((((.((((...)))).))))......)))))).........)))))) ( -28.90) >consensus GAUGCA_______CAUCCAUUGCAU______________CCAGUGCAACGAAGUGCAACGCCAUCAAUUUUUCCCACAGCCGAGGGCUCUGGGUUAU________AUUUAACAAAUGCAU ....................(((((.................((((......))))................((((.((((...)))).)))).....................))))). (-18.44 = -18.28 + -0.16)

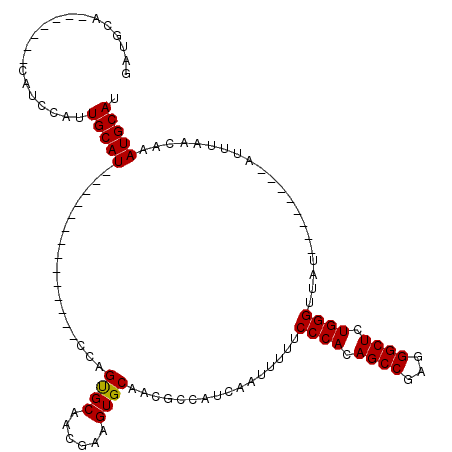
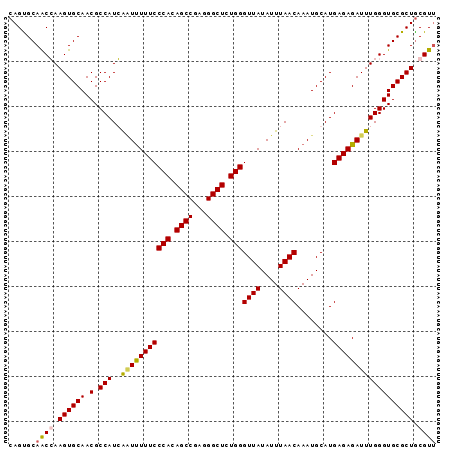
| Location | 9,077,291 – 9,077,382 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.65 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.56 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9077291 91 - 20766785 AUGCAUUUGUUAAAU--------AUAACCCAGAGCCCUCGGCUGUGGGAAAAAAUGAUGGCGUUGCACUUCGUUGCACUGG--------------AUGCAAUGGAUG-------UGCAUC ((((..((((.....--------))))((((.((((...)))).))))...........))))((((((((((((((....--------------.))))))))).)-------)))).. ( -32.40) >DroSec_CAF1 42207 89 - 1 AUGCAUUUGUUAAAU--------AUAACCCAGAGCCCUCGGCUGUGGGAAAGAUCCAUGGCGUUGCACUUGGUUGCACUGG--------------AUGCAACGAAUG-------U--AAC .((((((((((....--------....((((.((((...)))).))))....(((((..(((...........)))..)))--------------))..))))))))-------)--).. ( -30.80) >DroSim_CAF1 44014 80 - 1 AUGCAUUUGUUAAAU--------AUAACCCAGAGCCCUCGGCUGUGGGAAAAAUCCAUGGCGUUGCACUUGGUUGCACUGG--------------AUGCAA------------------C .(((...........--------....((((.((((...)))).))))....(((((..(((...........)))..)))--------------))))).------------------. ( -24.00) >DroEre_CAF1 49575 98 - 1 AUGCAUUUGUUAAAU--------AUAACCCAGAGCCCUCGGCUGUGGGAAAAACUGAUGGCGUUGCACUUCGCUGCGCUGG--------------AUGCAUCGGAUGCAUUGGAUGCAUU ((((((((.......--------....((((.((((...)))).))))..........(((((.((.....)).))))).(--------------((((((...))))))))))))))). ( -34.90) >DroYak_CAF1 45356 120 - 1 AUGCAUUUGAUAAAUAUACAUAUAUAACCCAGAGCCCUCGGCUGUGGGAAAAACUGAUGGCGUUGCACUUCGCUGCACUGGAUGGGAUGCAGUGGAUGCAUUGGAUGCAUUGGAUGCAUU (((((((..((................((((.((((...)))).))))...........((((((((.(((((((((((.....)).)))))))))))))....))))))..))))))). ( -44.20) >consensus AUGCAUUUGUUAAAU________AUAACCCAGAGCCCUCGGCUGUGGGAAAAACUGAUGGCGUUGCACUUCGUUGCACUGG______________AUGCAACGGAUG_______UGCAUC .(((((.....................((((.((((...)))).))))............((.((((......)))).))...............))))).................... (-21.72 = -21.56 + -0.16)

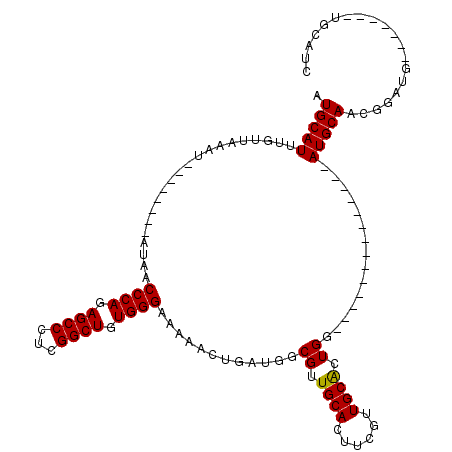
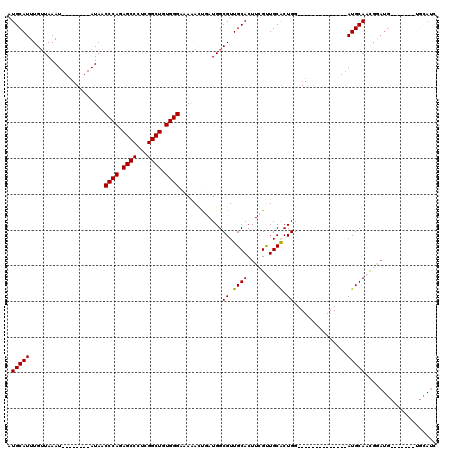
| Location | 9,077,310 – 9,077,405 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -27.98 |
| Energy contribution | -28.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9077310 95 + 20766785 CAGUGCAACGAAGUGCAACGCCAUCAUUUUUUCCCACAGCCGAGGGCUCUGGGUUAUAUUUAACAAAUGCAUGAGAGAUUUGGGUGCGCUACGUU ......((((.((((((.(.(((....(((((((((.((((...)))).)))((((....))))........))))))..)))))))))).)))) ( -28.40) >DroSec_CAF1 42224 95 + 1 CAGUGCAACCAAGUGCAACGCCAUGGAUCUUUCCCACAGCCGAGGGCUCUGGGUUAUAUUUAACAAAUGCAUGAGAGAUUUGGGUGCGCUGCGUU (((((((.(((.(((......))).(((((((((((.((((...)))).)))((((....))))........))))))))))).))))))).... ( -30.90) >DroSim_CAF1 44022 95 + 1 CAGUGCAACCAAGUGCAACGCCAUGGAUUUUUCCCACAGCCGAGGGCUCUGGGUUAUAUUUAACAAAUGCAUGAGAGAUUUGGGUGCGCUGCGUU (((((((.((((((.(....(((((.((((...(((.((((...)))).)))((((....)))))))).)))).).))))))).))))))).... ( -31.60) >DroEre_CAF1 49601 94 + 1 CAGCGCAGCGAAGUGCAACGCCAUCAGUUUUUCCCACAGCCGAGGGCUCUGGGUUAUAUUUAACAAAUGCAUGAGAGAUUUGGGUGCGCUGCGU- ..((((((((........((((...(((((((((((.((((...)))).)))((((....))))........))))))))..))))))))))))- ( -33.10) >consensus CAGUGCAACCAAGUGCAACGCCAUCAAUUUUUCCCACAGCCGAGGGCUCUGGGUUAUAUUUAACAAAUGCAUGAGAGAUUUGGGUGCGCUGCGUU ......((((.((((((.(.(((..(((((((((((.((((...)))).)))((((....))))........)))))))))))))))))).)))) (-27.98 = -28.23 + 0.25)

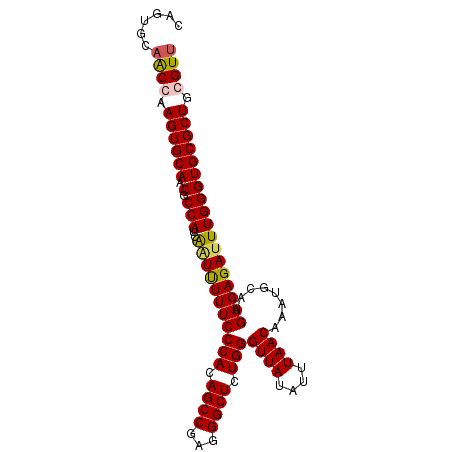
| Location | 9,077,310 – 9,077,405 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -28.09 |
| Energy contribution | -27.90 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.70 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9077310 95 - 20766785 AACGUAGCGCACCCAAAUCUCUCAUGCAUUUGUUAAAUAUAACCCAGAGCCCUCGGCUGUGGGAAAAAAUGAUGGCGUUGCACUUCGUUGCACUG ...(((((((...((((((......).)))))..........((((.((((...)))).))))...........))))))).............. ( -26.40) >DroSec_CAF1 42224 95 - 1 AACGCAGCGCACCCAAAUCUCUCAUGCAUUUGUUAAAUAUAACCCAGAGCCCUCGGCUGUGGGAAAGAUCCAUGGCGUUGCACUUGGUUGCACUG ...(((((((...((((((......).)))))..........((((.((((...)))).))))...........))))))).............. ( -28.40) >DroSim_CAF1 44022 95 - 1 AACGCAGCGCACCCAAAUCUCUCAUGCAUUUGUUAAAUAUAACCCAGAGCCCUCGGCUGUGGGAAAAAUCCAUGGCGUUGCACUUGGUUGCACUG ...(((((((...((((((......).)))))..........((((.((((...)))).))))...........))))))).............. ( -28.40) >DroEre_CAF1 49601 94 - 1 -ACGCAGCGCACCCAAAUCUCUCAUGCAUUUGUUAAAUAUAACCCAGAGCCCUCGGCUGUGGGAAAAACUGAUGGCGUUGCACUUCGCUGCGCUG -...((((((...((((((......).)))))..........((((.((((...)))).))))..........((((........)))))))))) ( -29.40) >consensus AACGCAGCGCACCCAAAUCUCUCAUGCAUUUGUUAAAUAUAACCCAGAGCCCUCGGCUGUGGGAAAAAUCCAUGGCGUUGCACUUCGUUGCACUG ...(((((((...((((((......).)))))..........((((.((((...)))).))))...........))))))).............. (-28.09 = -27.90 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:09 2006