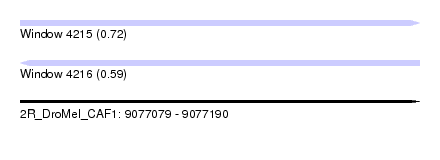
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,077,079 – 9,077,190 |
| Length | 111 |
| Max. P | 0.718196 |

| Location | 9,077,079 – 9,077,190 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -29.10 |
| Energy contribution | -30.44 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9077079 111 + 20766785 UGACAUACUGACCGCAGAGAGUGUACUUGAGCAAG-GGACUUCCCCGUGAAUCUGCCCGCCUGCUCAAGUGCUACGUAAUUUGCAUGGUCUGGAUUUUUUCAAAUUACAUGA-------- ...(((...(((((((((..(((((((((((((.(-((.....)))(((........))).))))))))))).))....)))))..))))((((....))))......))).-------- ( -36.10) >DroEre_CAF1 49346 116 + 1 UGGCAUACUGACCGCAGGGAGUGUACUUGAGCAGGG--GCUUCCCCGUGAAUCUGUCCGCCUGCUCAAGUGCUACACA-U-GGCAUGGUCUGGAUUUUGUCAAAUUACGCGAUGCUAUGA .......(((....)))...((((((((((((((((--(....)))(((........))))))))))))))).)).((-(-(((((.((...(((((....)))))..)).)))))))). ( -42.40) >DroYak_CAF1 45133 117 + 1 UGGCAUACUGACCGCAGGCAGUGUACUUGAGCUGGG--GCUUCCCCGUGAAGCUGUUCGCCUGCUCAAGUGCUACACA-UUUGCAUGGUCUGGAUUUUGUCGAAUUACGCGAUGCCAUGA ((((...(.(((((((((..((((((((((((.(((--(...))))(((((....)))))..)))))))))).))...-)))))..)))).)......((((.......))))))))... ( -42.70) >consensus UGGCAUACUGACCGCAGGGAGUGUACUUGAGCAGGG__GCUUCCCCGUGAAUCUGUCCGCCUGCUCAAGUGCUACACA_UUUGCAUGGUCUGGAUUUUGUCAAAUUACGCGAUGC_AUGA (((((..(.(((((((((..(((((((((((((((...((..............))...))))))))))))).))....)))))..)))).).....))))).................. (-29.10 = -30.44 + 1.34)

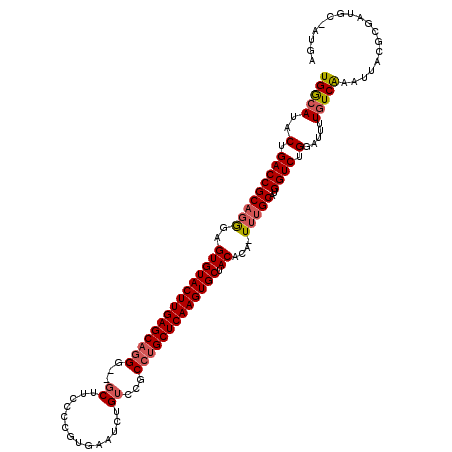
| Location | 9,077,079 – 9,077,190 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -25.67 |
| Energy contribution | -26.45 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9077079 111 - 20766785 --------UCAUGUAAUUUGAAAAAAUCCAGACCAUGCAAAUUACGUAGCACUUGAGCAGGCGGGCAGAUUCACGGGGAAGUCC-CUUGCUCAAGUACACUCUCUGCGGUCAGUAUGUCA --------.......((((....))))...(((.((((......(((((.(((((((((((.((((...(((.....)))))))-))))))))))).......)))))....))))))). ( -31.30) >DroEre_CAF1 49346 116 - 1 UCAUAGCAUCGCGUAAUUUGACAAAAUCCAGACCAUGCC-A-UGUGUAGCACUUGAGCAGGCGGACAGAUUCACGGGGAAGC--CCCUGCUCAAGUACACUCCCUGCGGUCAGUAUGCCA .....((((.((((......))........((((..((.-.-.((((...(((((((((((.((.....(((.....))).)--)))))))))))))))).....)))))).)))))).. ( -36.20) >DroYak_CAF1 45133 117 - 1 UCAUGGCAUCGCGUAAUUCGACAAAAUCCAGACCAUGCAAA-UGUGUAGCACUUGAGCAGGCGAACAGCUUCACGGGGAAGC--CCCAGCUCAAGUACACUGCCUGCGGUCAGUAUGCCA ...((((((.((((......))........((((..(((..-.((((...((((((((((((.....))))...((((...)--))).))))))))))))....))))))).)))))))) ( -41.60) >consensus UCAU_GCAUCGCGUAAUUUGACAAAAUCCAGACCAUGCAAA_UGUGUAGCACUUGAGCAGGCGGACAGAUUCACGGGGAAGC__CCCUGCUCAAGUACACUCCCUGCGGUCAGUAUGCCA ..........(((((((((....))))...((((..(((....((((...(((((((((((........(((.....))).....)))))))))))))))....)))))))..))))).. (-25.67 = -26.45 + 0.78)

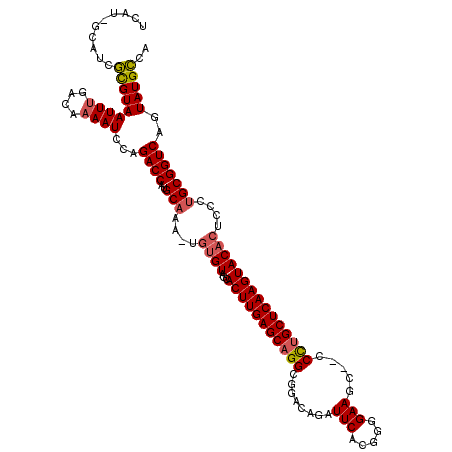
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:05 2006