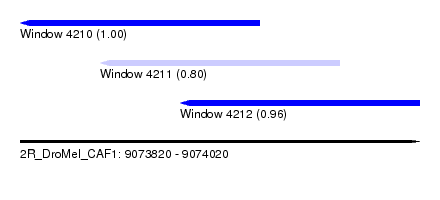
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,073,820 – 9,074,020 |
| Length | 200 |
| Max. P | 0.998014 |

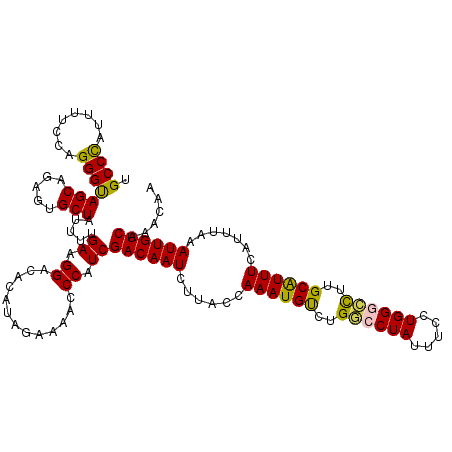
| Location | 9,073,820 – 9,073,940 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -26.30 |
| Energy contribution | -25.62 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9073820 120 - 20766785 UGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACAUAUAGAAAACCCAUCGACAAUCUUACCAAAUGUCUGGCCUAUUUCGUGGGCCUUGCAUUUCCUUUAAAUUGUCAAACAA ......((((.(((((((((.....))))))))).)))).................((((((......((((((..(((((((...)))))))..)))))).......))))))...... ( -30.62) >DroSec_CAF1 38766 120 - 1 UGCCUAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACACAUAGUAAACCCAUCGACAAUCUUACCAAAUGUCUGGCCUAUUUCCUGGACCUUGCAUUUCUUUUAAAUUGUCAAACAA (((((......(((((((((.....))))))))).))).))...............((((((......((((((..((((........)).))..)))))).......))))))...... ( -25.02) >DroSim_CAF1 40549 120 - 1 UGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACACAUAGAAAACCCAUCGACAAUCUUACCAAAUGUCUGGCCUAUUUCCUGGGCCUUGCAUUUCUUUUAAAUUGUCAAACAA ......((((((((((((((.....)))))))))..(....)...)))))......((((((......((((((..((((((.....))))))..)))))).......))))))...... ( -30.42) >DroEre_CAF1 44452 120 - 1 UGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACACAUAGCAAACCCAUCGACAAUCUUACCAAAUGCUUGACCUAAUUCCUGGUCUUUGCGUUUCAUUUAAAUUGUCAAACAA (((((........)))))......((((((..((.....)).))))))........((((((......((((((..((((........))))...)))))).......))))))...... ( -28.12) >DroYak_CAF1 40904 120 - 1 UGCCCACUUUCCAGGGCAGCAGAGUGCUAUUUCGAAGGACAUAUAGAAAACCCAUCGACAAUCUUACCAAAUGCUUGACCUAAUUCCUGGGUUUUGCGUUUCAUUUAAAUUGUCAAACAA (((((........)))))...(.((.((((.((....))...))))...)).)...((((((......((((((..((((((.....))))))..)))))).......))))))...... ( -31.42) >consensus UGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACACAUAGAAAACCCAUCGACAAUCUUACCAAAUGUCUGGCCUAUUUCCUGGGCCUUGCAUUUCAUUUAAAUUGUCAAACAA .((((........))))(((.....))).....((.((.............)).))((((((......((((((..((((((.....))))))..)))))).......))))))...... (-26.30 = -25.62 + -0.68)

| Location | 9,073,860 – 9,073,980 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -24.20 |
| Energy contribution | -23.60 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9073860 120 - 20766785 UUAUCUUUCAGAACCACGUGUUGUCCAAUGCACGCUCAACUGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACAUAUAGAAAACCCAUCGACAAUCUUACCAAAUGUCUGGCC ...((((((((((....((((........))))((((..((((((........))))))..))))....))))))))))............(((..((((...........))))))).. ( -31.00) >DroSec_CAF1 38806 120 - 1 UUAUCUUUCAGAACCACAUGUUGUCCAAUGCACGCUCAACUGCCUAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACACAUAGUAAACCCAUCGACAAUCUUACCAAAUGUCUGGCC ...((((((((((.....(((........)))(((((..((((((........))))))..)))))...))))))))))............(((..((((...........))))))).. ( -26.70) >DroSim_CAF1 40589 120 - 1 UUAUCUUUCAGAACCACGUGUUGUCCAAUGCACGCUCAACUGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACACAUAGAAAACCCAUCGACAAUCUUACCAAAUGUCUGGCC ...((((((((((....((((........))))((((..((((((........))))))..))))....))))))))))............(((..((((...........))))))).. ( -31.00) >DroEre_CAF1 44492 120 - 1 UCAUUUUCUGGAACCACGUGUUGUCCUAUGCACGCUCUACUGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACACAUAGCAAACCCAUCGACAAUCUUACCAAAUGCUUGACC ........(((......((((.(((((...((((((((.((((((........)))))).))))))......)).))))))))).......))).......................... ( -28.02) >DroYak_CAF1 40944 120 - 1 UCAUUUUCUGGAACCACGUGUUGUCCUAUGCACGCUCGGCUGCCCACUUUCCAGGGCAGCAGAGUGCUAUUUCGAAGGACAUAUAGAAAACCCAUCGACAAUCUUACCAAAUGCUUGACC ...(((((((...........((((((.((..(((((.(((((((........))))))).)))))......)).))))))..))))))).............................. ( -32.12) >consensus UUAUCUUUCAGAACCACGUGUUGUCCAAUGCACGCUCAACUGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGACACAUAGAAAACCCAUCGACAAUCUUACCAAAUGUCUGGCC ...((((((((((....((((........))))((((..((((((........))))))..))))....))))))))))......................................... (-24.20 = -23.60 + -0.60)

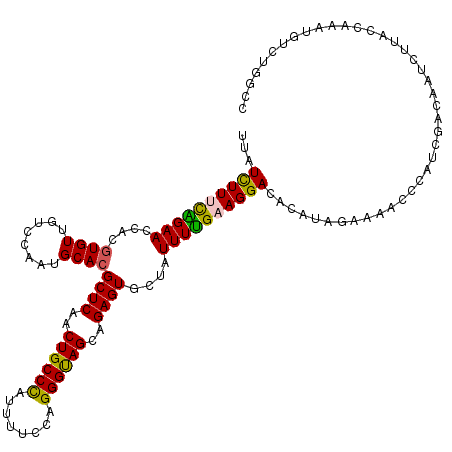
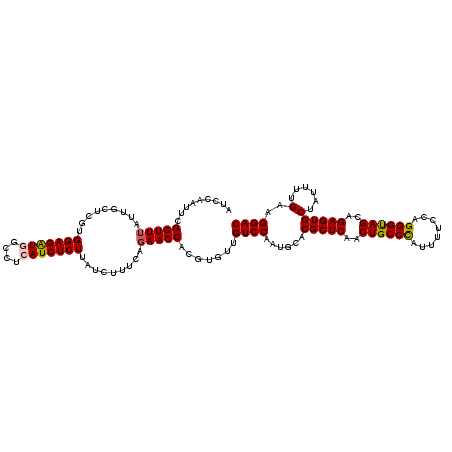
| Location | 9,073,900 – 9,074,020 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -35.81 |
| Consensus MFE | -30.65 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9073900 120 - 20766785 AUCCAAUUCGGUUUAUUGCUCGUGGAGAUGGCCUCAUCUCUUAUCUUUCAGAACCACGUGUUGUCCAAUGCACGCUCAACUGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGAC ........((((.....)).)).(((((((....)))))))..((((((((((....((((........))))((((..((((((........))))))..))))....)))))))))). ( -35.70) >DroSec_CAF1 38846 120 - 1 AUCAAAUCCGGUUAAUAACUCGUGGAGAUGGCCUCAUCUCUUAUCUUUCAGAACCACAUGUUGUCCAAUGCACGCUCAACUGCCUAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGAC ......(((((((..........(((((((....)))))))..........))))..........(((....(((((..((((((........))))))..))))).....)))..))). ( -31.35) >DroSim_CAF1 40629 120 - 1 AUCCAAUCCGGUUUAUUGCUCGUGGAGAUGGCCUCAUCUCUUAUCUUUCAGAACCACGUGUUGUCCAAUGCACGCUCAACUGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGAC ........((((.....)).)).(((((((....)))))))..((((((((((....((((........))))((((..((((((........))))))..))))....)))))))))). ( -35.90) >DroEre_CAF1 44532 120 - 1 AUCCAAAUCGGUUUAUUUCUCGUGGAGAUGGUUGAAUCUCUCAUUUUCUGGAACCACGUGUUGUCCUAUGCACGCUCUACUGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGAC .(((.....(((((.......((((((((......))))).)))......)))))..((((........))))(((((.((((((........)))))).)))))...........))). ( -36.62) >DroYak_CAF1 40984 120 - 1 AUUCAAUUCGGUUUAUUUUUGGUGGAGGUCGUUGCAUCUCUCAUUUUCUGGAACCACGUGUUGUCCUAUGCACGCUCGGCUGCCCACUUUCCAGGGCAGCAGAGUGCUAUUUCGAAGGAC .........(((((......(((((((((......))))).)))).....))))).......(((((.((..(((((.(((((((........))))))).)))))......)).))))) ( -39.50) >consensus AUCCAAUUCGGUUUAUUGCUCGUGGAGAUGGCCUCAUCUCUUAUCUUUCAGAACCACGUGUUGUCCAAUGCACGCUCAACUGCCCAUUUUCCAGGGUAGCAGAGUGCUAUUUUGAAGGAC .........(((((.........(((((((....))))))).........))))).......((((......(((((..((((((........))))))..)))))(......)..)))) (-30.65 = -30.77 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:01 2006