| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,073,628 – 9,073,780 |
| Length | 152 |
| Max. P | 0.997762 |

| Location | 9,073,628 – 9,073,748 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9073628 120 - 20766785 CAUGCACGCCCAUUUGCCCAUUUUCCAGGGUAAUUGGGUGCUAUUUUUGAAGGACAUAUAGAAAUCAUGUGAACAAUCUUACCGAAACGUUUUGCUAAUUUUUUGGCACUGCUUUUCAUU ......((((((.((((((........)))))).))))))...........(..((((((....).)))))..).........((((.((..((((((....))))))..)).))))... ( -33.20) >DroSec_CAF1 38609 120 - 1 UAUGCACGCCCAUUCGCCCAUUUUCUAGGGUAUAUGGGUGCUAUUUUUAAAGGACAUAUAGAAAUUAUGUGAACAAUCUUACCGAAACGUUUUGCUAACAUCUUGGCACAGCUUUUCAUU ......(((((((..((((........))))..)))))))...........(..(((((((...)))))))..).........((((.(((.((((((....)))))).))).))))... ( -32.00) >DroSim_CAF1 40366 120 - 1 UAUGCACGCCCAUUCCCCCAUUUUCCAGGGUAAUUGGGUGCUAUUUUUGAAGGGCAUAUAGAAAUUAUGUGAACAAUCUUACCGAAACGUUUUGCUAACUUCUUGGCACUGCUUUUCAUU ......((((((.(..(((........)))..).))))))...........(..(((((((...)))))))..).........((((.((..((((((....))))))..)).))))... ( -29.60) >DroEre_CAF1 44292 120 - 1 UAUGCACGCCCAUUUGCCCAUUUUCCAGGGUAAUUGGGUGCUAUUUUUCAAGGACAUAUAGAAAUCUUGUGAACAAUCUUACCGAAACGUUUUGCUAACUUCCUGGCACUGCUUUUCAUU ......((((((.((((((........)))))).))))))...........(..((((.((....))))))..).........((((.((..(((((......)))))..)).))))... ( -31.50) >DroYak_CAF1 40734 117 - 1 UAUGCACGCCCAUUUGCCCAUUUUACAGGGUAAUAGGGUGCUAUUUUUCAAGGACAUAUAGAAAUCUU---AACAAUCUUACCGAAACGUUGUGCUUAUUUUCAGGCACUGCUUUUCAUU ......(((((..((((((........))))))..)))))..........((((.....((....)).---.....))))...((((.((.((((((......)))))).)).))))... ( -28.60) >consensus UAUGCACGCCCAUUUGCCCAUUUUCCAGGGUAAUUGGGUGCUAUUUUUCAAGGACAUAUAGAAAUCAUGUGAACAAUCUUACCGAAACGUUUUGCUAACUUCUUGGCACUGCUUUUCAUU ......((((((.((((((........)))))).))))))...........................................((((.((..(((((......)))))..)).))))... (-25.94 = -26.62 + 0.68)

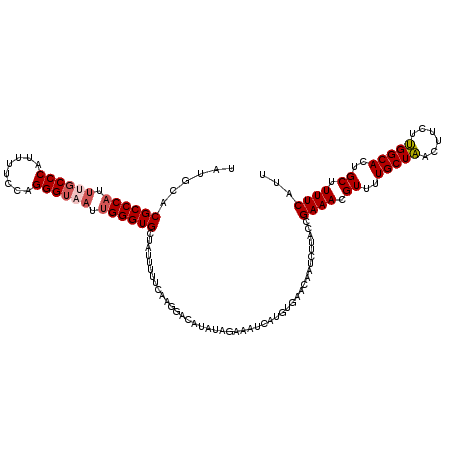
| Location | 9,073,668 – 9,073,780 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.08 |
| Mean single sequence MFE | -23.64 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9073668 112 + 20766785 AAGAUUGUUCACAUGAUUUCUAUAUGUCCUUCAAAAAUAGCACCCAAUUACCCUGGAAAAUGGGCAAAUGGGCGUGCAUGGGACAACGCACAUUUAAUU----AACCAAU----UUAUAU .((((((.((....))........((((((.........((.((((.((.(((........))).)).)))).))....))))))..............----...))))----)).... ( -22.22) >DroSec_CAF1 38649 111 + 1 AAGAUUGUUCACAUAAUUUCUAUAUGUCCUUUAAAAAUAGCACCCAUAUACCCUAGAAAAUGGGCGAAUGGGCGUGCAUAGGACAACGCAGAUGUACUU----AACUUAU----U-AUAU .(((((((....))))))).....((((((.........((.(((((...(((........)))...))))).))....))))))..............----.......----.-.... ( -21.52) >DroSim_CAF1 40406 111 + 1 AAGAUUGUUCACAUAAUUUCUAUAUGCCCUUCAAAAAUAGCACCCAAUUACCCUGGAAAAUGGGGGAAUGGGCGUGCAUAGGACAACGCAGAUGUACUU----AACUAAA----U-AUAU .(((((((....))))))).((((((((((((..........((((.((.(....).)).)))).))).))))((((((..(......)..))))))..----......)----)-))). ( -21.90) >DroEre_CAF1 44332 111 + 1 AAGAUUGUUCACAAGAUUUCUAUAUGUCCUUGAAAAAUAGCACCCAAUUACCCUGGAAAAUGGGCAAAUGGGCGUGCAUAGGACAACCCGGAAUUACUU----UAUUAAU----U-AUAU ..(((((.......((((((....((((((((.......((.((((.((.(((........))).)).)))).)).)).))))))....))))))....----...))))----)-.... ( -24.54) >DroYak_CAF1 40774 116 + 1 AAGAUUGUU---AAGAUUUCUAUAUGUCCUUGAAAAAUAGCACCCUAUUACCCUGUAAAAUGGGCAAAUGGGCGUGCAUAGGACAACACGCAAUAACUUAUUUCAAUAAUCAUGU-AUAU ..(((((((---.((....))...(((((((....))..((((((((((.(((........)))..)))))).))))...)))))...................)))))))....-.... ( -28.00) >consensus AAGAUUGUUCACAUGAUUUCUAUAUGUCCUUCAAAAAUAGCACCCAAUUACCCUGGAAAAUGGGCAAAUGGGCGUGCAUAGGACAACGCAGAUUUACUU____AACUAAU____U_AUAU ........................((((((.........((.((((.((.(((........))).)).)))).))....))))))................................... (-18.88 = -19.08 + 0.20)


| Location | 9,073,668 – 9,073,780 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.08 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -21.06 |
| Energy contribution | -22.10 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9073668 112 - 20766785 AUAUAA----AUUGGUU----AAUUAAAUGUGCGUUGUCCCAUGCACGCCCAUUUGCCCAUUUUCCAGGGUAAUUGGGUGCUAUUUUUGAAGGACAUAUAGAAAUCAUGUGAACAAUCUU ...(((----(.((((.----........((((((......))))))(((((.((((((........)))))).)))))))))..))))..(..((((((....).)))))..)...... ( -28.80) >DroSec_CAF1 38649 111 - 1 AUAU-A----AUAAGUU----AAGUACAUCUGCGUUGUCCUAUGCACGCCCAUUCGCCCAUUUUCUAGGGUAUAUGGGUGCUAUUUUUAAAGGACAUAUAGAAAUUAUGUGAACAAUCUU ((((-(----((..((.----....)).((((...((((((.....(((((((..((((........))))..))))))).((....)).))))))..)))).))))))).......... ( -28.20) >DroSim_CAF1 40406 111 - 1 AUAU-A----UUUAGUU----AAGUACAUCUGCGUUGUCCUAUGCACGCCCAUUCCCCCAUUUUCCAGGGUAAUUGGGUGCUAUUUUUGAAGGGCAUAUAGAAAUUAUGUGAACAAUCUU ....-.----....(((----..(((..((((...((((((...((((((((.(..(((........)))..).)))))).......)).))))))..))))...)))...)))...... ( -24.71) >DroEre_CAF1 44332 111 - 1 AUAU-A----AUUAAUA----AAGUAAUUCCGGGUUGUCCUAUGCACGCCCAUUUGCCCAUUUUCCAGGGUAAUUGGGUGCUAUUUUUCAAGGACAUAUAGAAAUCUUGUGAACAAUCUU ....-.----.......----......(((((((.((((((.....((((((.((((((........)))))).))))))..........)))))).........)))).)))....... ( -26.56) >DroYak_CAF1 40774 116 - 1 AUAU-ACAUGAUUAUUGAAAUAAGUUAUUGCGUGUUGUCCUAUGCACGCCCAUUUGCCCAUUUUACAGGGUAAUAGGGUGCUAUUUUUCAAGGACAUAUAGAAAUCUU---AACAAUCUU ....-..(((.((.((((((........((((((......))))))(((((..((((((........))))))..))))).....)))))).)))))...........---......... ( -25.40) >consensus AUAU_A____AUUAGUU____AAGUAAAUCUGCGUUGUCCUAUGCACGCCCAUUUGCCCAUUUUCCAGGGUAAUUGGGUGCUAUUUUUCAAGGACAUAUAGAAAUCAUGUGAACAAUCUU ...................................((((((.....((((((.((((((........)))))).))))))..........))))))........................ (-21.06 = -22.10 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:58 2006