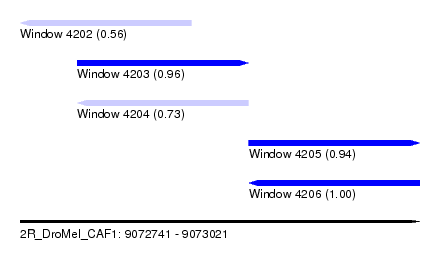
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,072,741 – 9,073,021 |
| Length | 280 |
| Max. P | 0.998794 |

| Location | 9,072,741 – 9,072,861 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -45.18 |
| Consensus MFE | -42.90 |
| Energy contribution | -42.82 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9072741 120 - 20766785 CCAUUUUGGCCAACUGCUGGAUGUACGGAGGCAAUGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGCGUGUUGAGAUGUGGCUUGGUCACGGGGGCAGGUGUGGUUG .......(((((.((((((((((..(((((((((..........))).))))))))))).((((..(((((((((...((((......))))))).)))))))))))))))...))))). ( -43.10) >DroSec_CAF1 37806 120 - 1 CCAUUUUGGCCAACUGCUGGAUGUGUGGAGGCAGGGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGCGUGUUGAGAUGUGGCUUGGUCACCGGAGCAGGUGUUGUUG .......(((((((..(.(((((((..(.((((((........)))))))..)))))))(((((((((..(.....)..)))).))))).)..).))))))(((......)))....... ( -40.90) >DroSim_CAF1 39457 120 - 1 CCAUUUUGGCCAACUGCUGAAUGUGUGGAGGCAGAGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGCGUGUUGAGAUGUGGCUUGGUCACGGGAGCAGGUGUUGUUG (((..(((..((..(((.((..(((((((((((((........)))).)))))))))..))..)))))..)))..)))((.((((....((((((...))))))..)))).))....... ( -39.90) >DroEre_CAF1 43394 120 - 1 CCAUUUUGGCCAACUGCUGGAUGUGCGGAGGCAGGGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGCGUGUUGAGAUGUGGCUUGGUCACAGGAGCAGGGGUGGUCG .......(((((.((((.(((((((.(..((((((........))))))..))))))))(((((..(((((((((...((((......))))))).)))))))))))))))...))))). ( -48.50) >DroYak_CAF1 39854 120 - 1 CCAUUUUGGCCAGCUGCUGGAUGUGCGGAGGCAGGGUUGAGUACUUGCCCUCCGCAUCCUCCUGCAUGAUCAGCUUGGGCGUGUUGAGAUGCGGCUUGGUCACAGGAGCAGGUGUGGUCG .......(((((.((((.(((((((.(..((((((........))))))..))))))))(((((..(((((((((...((((......))))))).)))))))))))))))...))))). ( -53.50) >consensus CCAUUUUGGCCAACUGCUGGAUGUGCGGAGGCAGGGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGCGUGUUGAGAUGUGGCUUGGUCACAGGAGCAGGUGUGGUUG .......(((((.((((.(((((((.((.((((((........)))))))).)))))))(((((..(((((((((...((((......))))))).)))))))))))))))...))))). (-42.90 = -42.82 + -0.08)

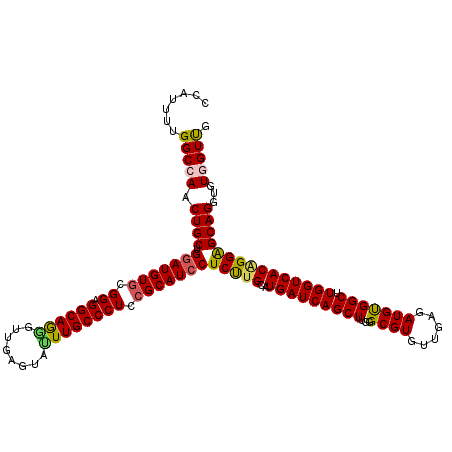
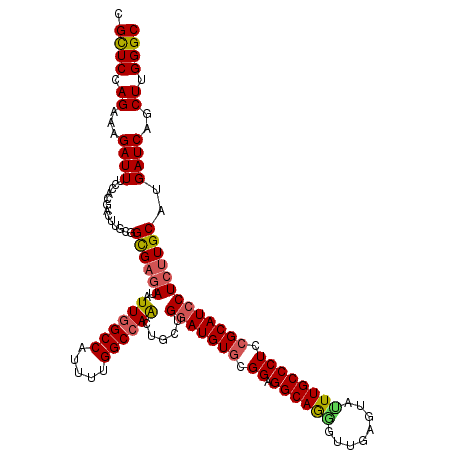
| Location | 9,072,781 – 9,072,901 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -34.34 |
| Energy contribution | -35.06 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9072781 120 + 20766785 GCCCAAGCUGAUCAUGCAAGAGGAUGCGGAGGGCAAAUACUCAACAUUGCCUCCGUACAUCCAGCAGUUGGCCAAAAUGGCCAAUAUUUCGCCGCAAGUCGUGGAAAUCUUUCUGGAGCG (((((.((.......))(((((((((((((((.(((..........)))))))))..)))))....(((((((.....))))))).(((((((....)..)))))).))))..))).)). ( -42.10) >DroSec_CAF1 37846 120 + 1 GCCCAAGCUGAUCAUGCAAGAGGAUGCGGAGGGCAAAUACUCAACCCUGCCUCCACACAUCCAGCAGUUGGCCAAAAUGGCCAAUAUCUCGCCCCAAGUCGUGGAAAUCUUUCUGGAGCG (((((....(((...((.((((((((.((..((((............))))..).).)))))....(((((((.....))))))).))).)).(((.....)))..)))....))).)). ( -37.60) >DroSim_CAF1 39497 120 + 1 GCCCAAGCUGAUCAUGCAAGAGGAUGCGGAGGGCAAAUACUCAACUCUGCCUCCACACAUUCAGCAGUUGGCCAAAAUGGCCAAUAUCUCGCCGCAAGUCGUGGAAAUCUUUCUGGAGCG (((((.(((((((......))(((.((((((.............)))))).)))......))))).(((((((.....)))))))......((((.....)))).........))).)). ( -41.12) >DroEre_CAF1 43434 120 + 1 GCCCAAGCUGAUCAUGCAAGAGGAUGCGGAGGGCAAAUACUCAACCCUGCCUCCGCACAUCCAGCAGUUGGCCAAAAUGGCUAAUAUCUCGCCCCAAGUUGUGGAAAUCUUUCUGGAACG ..(((((((......((.((((((((((((((.((............))))))))))..)))....(((((((.....))))))).))).))....)))).)))...(((....)))... ( -36.20) >DroYak_CAF1 39894 120 + 1 GCCCAAGCUGAUCAUGCAGGAGGAUGCGGAGGGCAAGUACUCAACCCUGCCUCCGCACAUCCAGCAGCUGGCCAAAAUGGCCAAUAUCUCACCGCAAGUCGUGGAAAUCUUUCUGGAGCG ((((((((((.((......))(((((((((((.((.((.....))..))))))))))..)))..)))))((((.....)))).........((((.....)))).........))).)). ( -43.00) >consensus GCCCAAGCUGAUCAUGCAAGAGGAUGCGGAGGGCAAAUACUCAACCCUGCCUCCGCACAUCCAGCAGUUGGCCAAAAUGGCCAAUAUCUCGCCGCAAGUCGUGGAAAUCUUUCUGGAGCG (((((.((.......))(((((((((((((((.((............))))))))..)))))....(((((((.....)))))))......((((.....))))...))))..))).)). (-34.34 = -35.06 + 0.72)

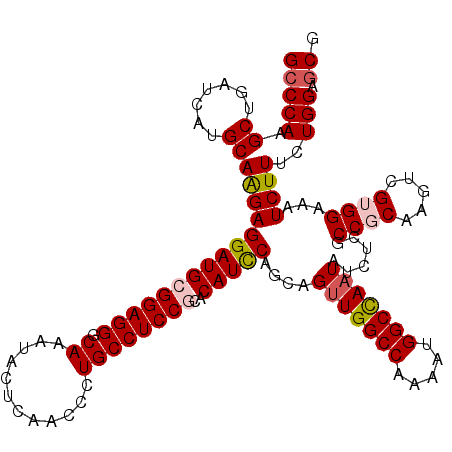
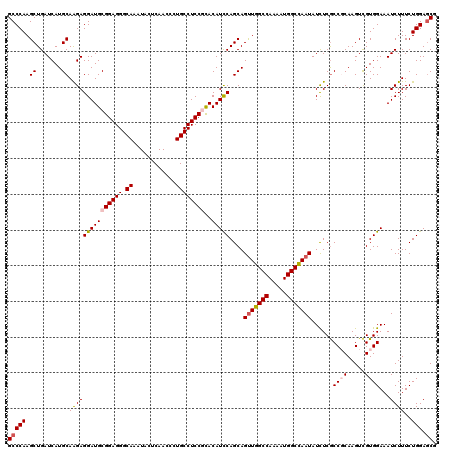
| Location | 9,072,781 – 9,072,901 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -42.79 |
| Consensus MFE | -37.46 |
| Energy contribution | -37.78 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9072781 120 - 20766785 CGCUCCAGAAAGAUUUCCACGACUUGCGGCGAAAUAUUGGCCAUUUUGGCCAACUGCUGGAUGUACGGAGGCAAUGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGC .((((.((...((((..((.((...((((.......((((((.....)))))))))).(((((..(((((((((..........))).))))))))))))).))...))))..)).)))) ( -38.71) >DroSec_CAF1 37846 120 - 1 CGCUCCAGAAAGAUUUCCACGACUUGGGGCGAGAUAUUGGCCAUUUUGGCCAACUGCUGGAUGUGUGGAGGCAGGGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGC .((((.((...((((((((.....))))((((((..((((((.....)))))).....(((((((..(.((((((........)))))))..)))))))))))))..))))..)).)))) ( -46.80) >DroSim_CAF1 39497 120 - 1 CGCUCCAGAAAGAUUUCCACGACUUGCGGCGAGAUAUUGGCCAUUUUGGCCAACUGCUGAAUGUGUGGAGGCAGAGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGC .((((.((...((((....((.....))((((((..((((((.....)))))).........(((((((((((((........)))).)))))))))..))))))..))))..)).)))) ( -41.80) >DroEre_CAF1 43434 120 - 1 CGUUCCAGAAAGAUUUCCACAACUUGGGGCGAGAUAUUAGCCAUUUUGGCCAACUGCUGGAUGUGCGGAGGCAGGGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGC ((((((((...............)))))))).........(((..(((..((..(((.(((.(((((((((((((........)))).)))))))))..))).)))))..)))..))).. ( -39.36) >DroYak_CAF1 39894 120 - 1 CGCUCCAGAAAGAUUUCCACGACUUGCGGUGAGAUAUUGGCCAUUUUGGCCAGCUGCUGGAUGUGCGGAGGCAGGGUUGAGUACUUGCCCUCCGCAUCCUCCUGCAUGAUCAGCUUGGGC .((((.((...(((((((((........))).))..((((((.....)))))).(((.(((.(((((((((((((........)))).)))))))))..))).))).))))..)).)))) ( -47.30) >consensus CGCUCCAGAAAGAUUUCCACGACUUGCGGCGAGAUAUUGGCCAUUUUGGCCAACUGCUGGAUGUGCGGAGGCAGGGUUGAGUAUUUGCCCUCCGCAUCCUCUUGCAUGAUCAGCUUGGGC .((((.((...((((.............((((((..((((((.....)))))).....(((((((.((.((((((........)))))))).)))))))))))))..))))..)).)))) (-37.46 = -37.78 + 0.32)

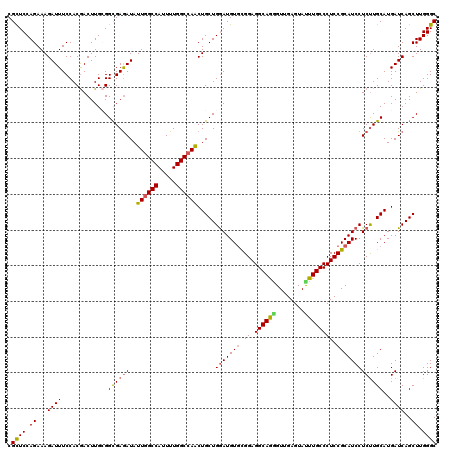
| Location | 9,072,901 – 9,073,021 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -48.52 |
| Consensus MFE | -42.38 |
| Energy contribution | -42.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9072901 120 + 20766785 GCAGCCCAAGUUGGCGGAACUGGCCAAGCGCCUGAGCACCCUGGCACUCAGUCCCGAACAGACCCAGGCCAUGGACACCCAGGUGCUCAAGGCCGUGCAGCACGCACUGAGCAACAACGA ...((.((..(((((.......)))))(.(((((((((((.((((.((..(((.......)))..))))))(((....)))))))))).)))))((((.....)))))).))........ ( -44.20) >DroSec_CAF1 37966 120 + 1 GCAACCCAAGCUUGCGGAACUUGCCAAGCGCCUGAGCACCCUGGCACUCAGUCCGGAGCAGACCCAGGCCAUGGACGCCCAGGUGCUCAAGGCCGUGCAGCACGCCCUGAGCAACAACGA .........(((((.((....(((...(((((((((((((..(((.....(((((..((........))..))))))))..))))))).))).)))...)))..)).)))))........ ( -44.90) >DroSim_CAF1 39617 120 + 1 GCAACCCAAGCUGGCGGAACUGGCCAAGCGCCUGAGCACCCUGGCACUCAGUCCGGAGCAGACCCAGGCCAUGGACGCCCAGGUGCUCAAGGCCGUGCAGCACGCCCUGAGCAACAACGA .........((((((((.((.((((.((((((((.((..((((((.((..(((.......)))..)))))).))..)).))))))))...))))))....).))))...)))........ ( -46.60) >DroEre_CAF1 43554 120 + 1 GCAGCCCAAGCUGGCCGAACUGGCCAAGCGCCUGAGCACCCUGGCCCUCAGUCCGGAGCAGACCCAGGCCAUGGGCAACCAGGUGCUCAGGGCCGUGCAGAACGCACUGCGCAACAACGA ...((((..(((((((.....)))).)))...((((((((.(((((....(((.......)))...)))))(((....))))))))))))))).((((((......))))))........ ( -57.70) >DroYak_CAF1 40014 120 + 1 GCAGCCCAAGCUGGCGGAACUGGCCAAGCGCUUGAGCACCCUGGCACUCAGUCCGGAGCAGACCCAGGCCAUGGACGCCCAGGUGCUCAAGGCCGUGCAGCACGCACUGAGCAGCAACGA ((.((.((.((((((.......))).))).((((((((((..(((.....(((((..((........))..))))))))..))))))))))...((((.....)))))).)).))..... ( -49.20) >consensus GCAGCCCAAGCUGGCGGAACUGGCCAAGCGCCUGAGCACCCUGGCACUCAGUCCGGAGCAGACCCAGGCCAUGGACGCCCAGGUGCUCAAGGCCGUGCAGCACGCACUGAGCAACAACGA ((.......((((((.......))).)))(((((((((((.((((.((..(((.......)))..))))))(((....)))))))))).)))).((((.....))))...))........ (-42.38 = -42.50 + 0.12)

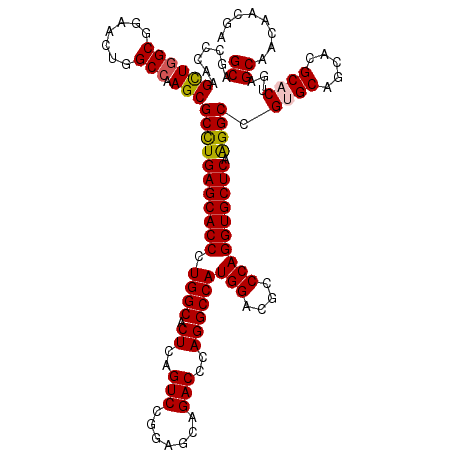
| Location | 9,072,901 – 9,073,021 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -59.98 |
| Consensus MFE | -54.56 |
| Energy contribution | -54.44 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9072901 120 - 20766785 UCGUUGUUGCUCAGUGCGUGCUGCACGGCCUUGAGCACCUGGGUGUCCAUGGCCUGGGUCUGUUCGGGACUGAGUGCCAGGGUGCUCAGGCGCUUGGCCAGUUCCGCCAACUUGGGCUGC .....((.(((((((((.....))))((((..((((.((((((..(((.((((((.(((((.....))))).)).)))))))..)))))).)))))))).............))))).)) ( -59.40) >DroSec_CAF1 37966 120 - 1 UCGUUGUUGCUCAGGGCGUGCUGCACGGCCUUGAGCACCUGGGCGUCCAUGGCCUGGGUCUGCUCCGGACUGAGUGCCAGGGUGCUCAGGCGCUUGGCAAGUUCCGCAAGCUUGGGUUGC .....((.(((((((.(.(((.(.((.(((..((((.(((((((..((.((((((.((((((...)))))).)).))))))..))))))).)))))))..)).).))).)))))))).)) ( -59.10) >DroSim_CAF1 39617 120 - 1 UCGUUGUUGCUCAGGGCGUGCUGCACGGCCUUGAGCACCUGGGCGUCCAUGGCCUGGGUCUGCUCCGGACUGAGUGCCAGGGUGCUCAGGCGCUUGGCCAGUUCCGCCAGCUUGGGUUGC .....((.(((((.((((.(((....((((..((((.(((((((..((.((((((.((((((...)))))).)).))))))..))))))).)))))))))))..))))....))))).)) ( -60.90) >DroEre_CAF1 43554 120 - 1 UCGUUGUUGCGCAGUGCGUUCUGCACGGCCCUGAGCACCUGGUUGCCCAUGGCCUGGGUCUGCUCCGGACUGAGGGCCAGGGUGCUCAGGCGCUUGGCCAGUUCGGCCAGCUUGGGCUGC ..........((((......)))).((((((.((((.(((((..((((.((((((.((((((...))))))..))))))))))..))))).))))((((.....)))).....)))))). ( -62.80) >DroYak_CAF1 40014 120 - 1 UCGUUGCUGCUCAGUGCGUGCUGCACGGCCUUGAGCACCUGGGCGUCCAUGGCCUGGGUCUGCUCCGGACUGAGUGCCAGGGUGCUCAAGCGCUUGGCCAGUUCCGCCAGCUUGGGCUGC .((..((((..((((((.....))))((((((((((((((((....)).((((((.((((((...)))))).)).))))))))))))))).)))))..))))..)).((((....)))). ( -57.70) >consensus UCGUUGUUGCUCAGUGCGUGCUGCACGGCCUUGAGCACCUGGGCGUCCAUGGCCUGGGUCUGCUCCGGACUGAGUGCCAGGGUGCUCAGGCGCUUGGCCAGUUCCGCCAGCUUGGGCUGC .....((.((((((.(((((..((..((((..((((.(((((((((((.((((((.(((((.....))))).)).))))))))))))))).)))))))).))..)))..)))))))).)) (-54.56 = -54.44 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:55 2006