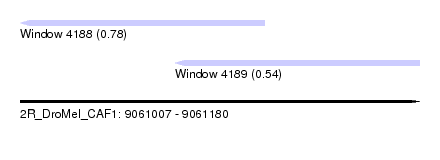
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,061,007 – 9,061,180 |
| Length | 173 |
| Max. P | 0.775864 |

| Location | 9,061,007 – 9,061,113 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -31.16 |
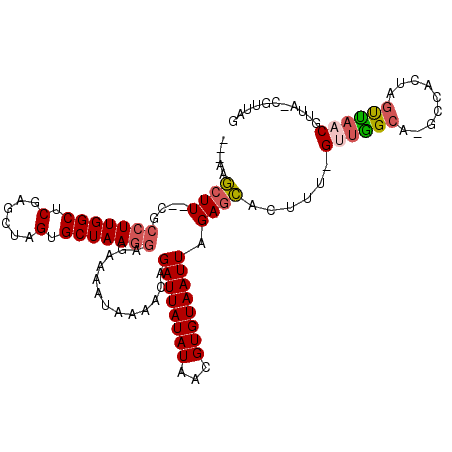
| Consensus MFE | -12.30 |
| Energy contribution | -14.74 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9061007 106 - 20766785 GCACUUU-GUUGGCAUGCCACUAGUUAACGUUAACGUUAACGUUAACGCUGUUUUCUAGCGCACACAUGACUUGGCAUUUCUGGGCUCCGAGCCGCAAGCCCACUUU .......-((.(((((((((..(((((..(((((((....)))))))((((.....)))).......)))))))))))...(((((.....))).)).))).))... ( -33.20) >DroSec_CAF1 26244 99 - 1 GCACUUU-GUUGGCA-GCCACUAGUUAACGUUAACGUUAG------CGUUGUUUUCUAGCGCACACAUGACUUGGCAUUUCUGGGCUCCGAGGCCCAAGCCCACUUU .......-((.(((.-((((..((((((((....)))).(------(((((.....))))))......)))))))).....((((((....)))))).))).))... ( -32.50) >DroSim_CAF1 28316 99 - 1 GCACUCU-GUUGGCA-GCCACUGGUUAACGUUAACGUUAG------CGUUGUUUUCUAGCGCACACAUGACUUGGCAUUUCUGGGCUCCGAGCCCCAAGCCCACUUU .......-((.(((.-((((..((((((((....)))).(------(((((.....))))))......))))))))......((((.....))))...))).))... ( -30.20) >DroYak_CAF1 28093 84 - 1 GCACUUU-GUUGGCA-GCCACUGGUCAACG---------------------UUUUCUAGCGCAGACAUGACUUGGCAUUUCUGGGCUCCAAGCCCCAAGCCCACUUU .......-((.(((.-((((..(((((..(---------------------(((.(....).)))).)))))))))......((((.....))))...))).))... ( -24.70) >DroAna_CAF1 30782 96 - 1 GUACUUUUGAUAGCGAGCCG--AGCUU-CAGUA-GCUUAG------CUGGCUUUUCUAGUGCAGACAUGACUUGGCAUUUGUGGGGUCUGGGCCCCAA-CCCACUUU (((((...((.(((.(((..--((((.-....)-)))..)------)).)))..)).)))))..........(((......((((((....)))))).-.))).... ( -35.20) >consensus GCACUUU_GUUGGCA_GCCACUAGUUAACGUUAACGUUAG______CGUUGUUUUCUAGCGCACACAUGACUUGGCAUUUCUGGGCUCCGAGCCCCAAGCCCACUUU ........((.(((..((((..(((((.......((((((...............))))))......))))))))).....((((........)))).))).))... (-12.30 = -14.74 + 2.44)

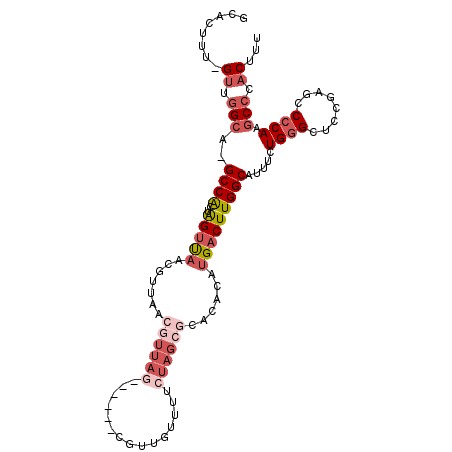
| Location | 9,061,074 – 9,061,180 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -17.54 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9061074 106 - 20766785 ---AAGCUU--CGCCUUGGCUCGAGCUAGUGCUAAGGAGAAAAUAAAACAGAUUAUAUAACGUGUAAUUAGAGCACUUU-GUUGGCAUGCCACUAGUUAACGUUAACGUUAA ---.(((((--(.(((((((.(......).))))))).)))................((((((.((((((((((.....-)))((....)).)))))))))))))..))).. ( -27.20) >DroSec_CAF1 26305 105 - 1 ---AAGCUU--CGCCUUGGCUCGAGCUAGUGCUAAGGAGAAAAUAAAACAGAUUAUAUAACGUGUAAUUAGAGCACUUU-GUUGGCA-GCCACUAGUUAACGUUAACGUUAG ---.(((((--(.(((((((.(......).))))))).)))................((((((.(((((((.((.((..-...))..-))..)))))))))))))..))).. ( -25.00) >DroSim_CAF1 28377 105 - 1 ---AAGCUU--CGCCUUGGCUCGAGCUAGUGCUAAGGAGAAAAUAAAACAGAUUAUAUAACGUGUAAUUAGAGCACUCU-GUUGGCA-GCCACUGGUUAACGUUAACGUUAG ---......--.(((.((((((.(((.((((((..(............).((((((((...))))))))..))))))..-))).).)-))))..)))(((((....))))). ( -26.90) >DroEre_CAF1 31969 95 - 1 ---AAGCUU--UGCCUUGGCUCGAGCUGGUGCUAAGGAGAAAAUAAAACAGAUUAUAUAACGUGUAAUUAGAGCACUUU-GCUGGCA-GCCACUGCUUAACG---------- ---......--.((..((((((.(((.((((((..(............).((((((((...))))))))..))))))..-))).).)-))))..))......---------- ( -25.00) >DroYak_CAF1 28149 95 - 1 ---GAGCUU--UACCUUGGCUCGAGCUGGUGCUAAGGAGAAAAUAAAACAGAUUAUAUAACGUGUAAUUAGAGCACUUU-GUUGGCA-GCCACUGGUCAACG---------- ---..((((--(.(((((((.(......).))))))).............((((((((...))))))))))))).....-((((((.-.......)))))).---------- ( -25.40) >DroAna_CAF1 30842 108 - 1 AAAAAAAUUCCAGCCUUGGCUCGCGUCUGUGCUAAAAAGAAAAUAAAAGCGAUUAUAUAACGUGUAAUUAGAGUACUUUUGAUAGCGAGCCG--AGCUU-CAGUA-GCUUAG ...........(((.((((((((((((.(((((.................((((((((...))))))))..)))))....))).))))))))--)))).-.....-...... ( -26.41) >consensus ___AAGCUU__CGCCUUGGCUCGAGCUAGUGCUAAGGAGAAAAUAAAACAGAUUAUAUAACGUGUAAUUAGAGCACUUU_GUUGGCA_GCCACUAGUUAACGUUA_CGUUAG .....((((....(((((((.(......).))))))).............((((((((...)))))))).))))......((((((.........))))))........... (-17.54 = -18.02 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:38 2006