
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,033,450 – 9,033,541 |
| Length | 91 |
| Max. P | 0.895392 |

| Location | 9,033,450 – 9,033,541 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.14 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -8.71 |
| Energy contribution | -8.93 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
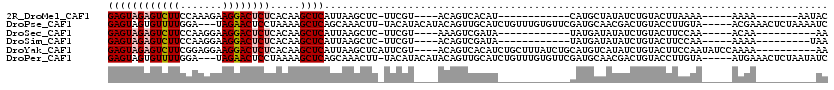
>2R_DroMel_CAF1 9033450 91 + 20766785 GAGUAGAGUCUUCCAAAGAAGGACUCUCACAAGCUCAUUAAGCUC-UUCGU----ACAGUCACAU------------CAUGCUAUAUCUGUACUUAAAA-----AAAA-------AAUAC ((((((((((((.......)))))))).....)))).........-...((----((((......------------..........))))))......-----....-------..... ( -17.79) >DroPse_CAF1 10054 111 + 1 GAGUAGUGUUUUGGA---UAGAACUCCUAAAAGCUCAGCAAACUU-UACAUACAUACAGUUGCAUCUGUUUGUGUUCGAUGCAACGACUGUACCUUGUA-----ACGAAACUCUAAAAUC ((((.(.((((((((---......)))..))))).)......(.(-((((....((((((((((((...........))))))...))))))...))))-----).)..))))....... ( -23.40) >DroSec_CAF1 9735 88 + 1 GAGUAGAGUCUUCCAAGGAAGGACUCUCACAAGCUCAUUAAGCUC-UUCGU----AAAGUCGAUA------------UAUGAUAUAUCUGUACUUCCAA-----ACAA----------AA (((((((((((((....))).)))))).....)))).........-.....----.(((((((((------------((...)))))).).))))....-----....----------.. ( -17.10) >DroSim_CAF1 9124 89 + 1 GAGUAGAGUCUUCCAAGGAAGGACUCUCACAAGCUCAUUAAGCUC-UUCGU----ACAGUCGAUA------------UAUGAUAUAUCUGUACUUCCAA-----AAAA---------UAA (((((((((((((....))).)))))).....)))).........-...((----((((...(((------------(...))))..))))))......-----....---------... ( -21.30) >DroYak_CAF1 9774 106 + 1 GAGUAGAGUCUUCGGAGGAAGGACUCUCACAAGCUCAUUAAGCUCAUUCGU----ACAGUCACAUCUGCUUUAUCUGCAUGUCAUAUCUGUACUUCCAAUAUCCAAAA----------AA (((((((((((((....))).))))))....((((.....)))).))))((----((((......((((.......))).)......))))))...............----------.. ( -22.70) >DroPer_CAF1 6813 111 + 1 GAGUAGUGUUUUGGA---UAGAACUCCUAAAAGCUCAGCAAACUU-UACAUACAUACAGUUGCAUCUGUUUGUGUUCGAUGCAACGACUGUACCUUGUA-----AUGAAACUCUAAUAUC ((((.(.((((((((---......)))..))))).)......(.(-((((....((((((((((((...........))))))...))))))...))))-----).)..))))....... ( -23.40) >consensus GAGUAGAGUCUUCCAAGGAAGGACUCUCACAAGCUCAUUAAGCUC_UUCGU____ACAGUCGCAU____________CAUGCUAUAUCUGUACUUCCAA_____AAAA__________AA ((((((((((((.......)))))))).....)))).................................................................................... ( -8.71 = -8.93 + 0.22)
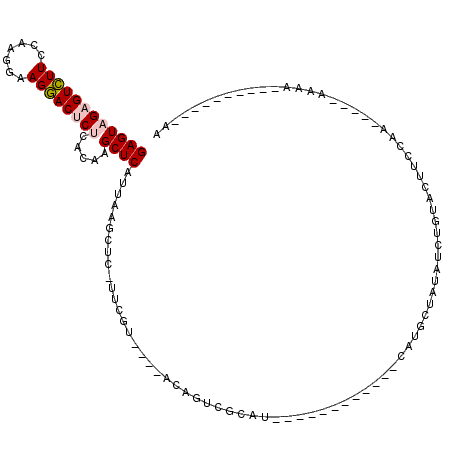
| Location | 9,033,450 – 9,033,541 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.14 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -9.51 |
| Energy contribution | -8.73 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9033450 91 - 20766785 GUAUU-------UUUU-----UUUUAAGUACAGAUAUAGCAUG------------AUGUGACUGU----ACGAA-GAGCUUAAUGAGCUUGUGAGAGUCCUUCUUUGGAAGACUCUACUC .....-------....-----......((((((.((((.....------------.)))).))))----))...-((((((...))))))((.((((((.(((....))))))))))).. ( -24.20) >DroPse_CAF1 10054 111 - 1 GAUUUUAGAGUUUCGU-----UACAAGGUACAGUCGUUGCAUCGAACACAAACAGAUGCAACUGUAUGUAUGUA-AAGUUUGCUGAGCUUUUAGGAGUUCUA---UCCAAAACACUACUC .......((((....(-----(((((.(((((((...((((((...........))))))))))))).).))))-).((((...((((((....))))))..---....))))...)))) ( -27.40) >DroSec_CAF1 9735 88 - 1 UU----------UUGU-----UUGGAAGUACAGAUAUAUCAUA------------UAUCGACUUU----ACGAA-GAGCUUAAUGAGCUUGUGAGAGUCCUUCCUUGGAAGACUCUACUC .(----------((((-----...(((((...((((((...))------------)))).)))))----)))))-((((((...))))))((.((((((.(((....))))))))))).. ( -23.90) >DroSim_CAF1 9124 89 - 1 UUA---------UUUU-----UUGGAAGUACAGAUAUAUCAUA------------UAUCGACUGU----ACGAA-GAGCUUAAUGAGCUUGUGAGAGUCCUUCCUUGGAAGACUCUACUC ...---------...(-----(((..(((...((((((...))------------)))).)))..----.))))-((((((...))))))((.((((((.(((....))))))))))).. ( -22.90) >DroYak_CAF1 9774 106 - 1 UU----------UUUUGGAUAUUGGAAGUACAGAUAUGACAUGCAGAUAAAGCAGAUGUGACUGU----ACGAAUGAGCUUAAUGAGCUUGUGAGAGUCCUUCCUCCGAAGACUCUACUC .(----------(((((((....(((.((((((.((((...(((.......)))..)))).))))----))..(..(((((...)))))..).....)))....))))))))........ ( -27.70) >DroPer_CAF1 6813 111 - 1 GAUAUUAGAGUUUCAU-----UACAAGGUACAGUCGUUGCAUCGAACACAAACAGAUGCAACUGUAUGUAUGUA-AAGUUUGCUGAGCUUUUAGGAGUUCUA---UCCAAAACACUACUC .......((((....(-----(((((.(((((((...((((((...........))))))))))))).).))))-).((((...((((((....))))))..---....))))...)))) ( -27.50) >consensus GU__________UUUU_____UUGGAAGUACAGAUAUAGCAUA____________AUGCGACUGU____ACGAA_GAGCUUAAUGAGCUUGUGAGAGUCCUUCCUUCGAAGACUCUACUC ..........................(((.........................................((...((((((...)))))).))((((((.(((....)))))))))))). ( -9.51 = -8.73 + -0.77)

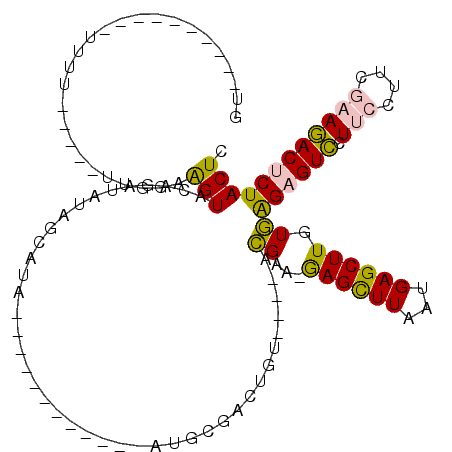
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:31 2006