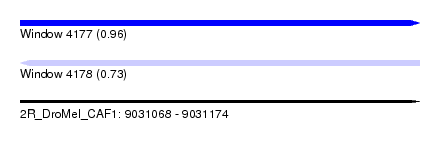
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,031,068 – 9,031,174 |
| Length | 106 |
| Max. P | 0.964259 |

| Location | 9,031,068 – 9,031,174 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 71.09 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -8.88 |
| Energy contribution | -11.63 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
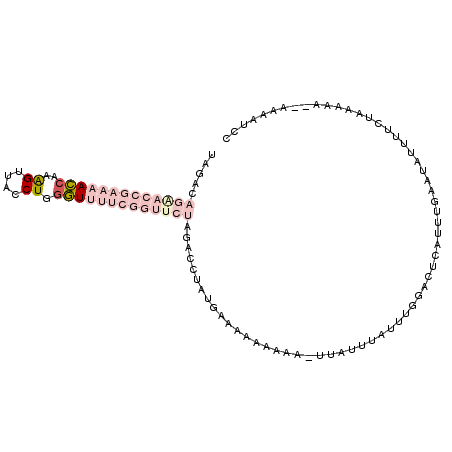
>2R_DroMel_CAF1 9031068 106 + 20766785 UAGACAGAACCGAAAACCAAAGUUACCUGGGUUUUCGGUUCUAGACCUAUGAAAAAAAAAUUUAUUUAUUUGGACUCAUUUGAAUAUAUUCUAAAAAAUAAAAUCC ((((.(((((((((((((..((....)).)))))))))))))....................((((((..((....))..))))))...))))............. ( -24.10) >DroPse_CAF1 7636 84 + 1 GAG------------AACAAGGGUGCCUGGUUUUUCGGUUUCACAUCAAUGAAACACA-----CUUUCUUUGGAGUUCUUUUAACAUUA----ACAA-AAAAUUUU (((------------(((....(((.((((....))))...))).((((.((((....-----.)))).)))).)))))).........----....-........ ( -12.40) >DroSec_CAF1 7394 102 + 1 UAGACAGAACCGAAAACCAAAGUUACCUGUGUUUUCGGUUCUAGACCUAUGAAAA-AAAA-UUAUUUAUUUGGACUCAUUUGAAUAUUUUCUAAAAA--AAAAUCC ((((.((((((((((((((........)).)))))))))))).............-....-.((((((..((....))..))))))...))))....--....... ( -22.30) >DroSim_CAF1 6744 102 + 1 UAGACAGGACCGAAAACCAAAGUUACCUGGGUUUUCGGUUCUAGACCUAUGAAAAAAAAA-UUAUUUAUUUGGACUCAUUUGAAUAUUUUCUAAAA---AAAAUCC ((((.(((((((((((((..((....)).)))))))))))))..................-.((((((..((....))..))))))...))))...---....... ( -23.60) >DroYak_CAF1 7380 99 + 1 UAGACAGAACCGAAAACCAAAGUUGCCUGGGUUUUCGGUUCUAUACCUAUGAAAA---AA-UUAUCUAUCUGGACUCAUUGGAAUAUUUUCUAAAAA---AAAACA ((((.(((((((((((((..((....)).)))))))))))))...((.((((...---..-..............)))).)).......))))....---...... ( -23.81) >DroPer_CAF1 4396 84 + 1 GAG------------AACAAGGGUGCCUGGUUUUUCGGUUUCACAUCAAUGAAACACA-----CUUUCUUUGGAGUUCUUUUAACAUUA----ACAA-AAAAUUUU (((------------(((....(((.((((....))))...))).((((.((((....-----.)))).)))).)))))).........----....-........ ( -12.40) >consensus UAGACAGAACCGAAAACCAAAGUUACCUGGGUUUUCGGUUCUAGACCUAUGAAAAAAAAA_UUAUUUAUUUGGACUCAUUUGAAUAUUUUCUAAAAA__AAAAUCC .....(((((((((((((..((....)).)))))))))))))................................................................ ( -8.88 = -11.63 + 2.75)

| Location | 9,031,068 – 9,031,174 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 71.09 |
| Mean single sequence MFE | -21.08 |
| Consensus MFE | -7.45 |
| Energy contribution | -9.28 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
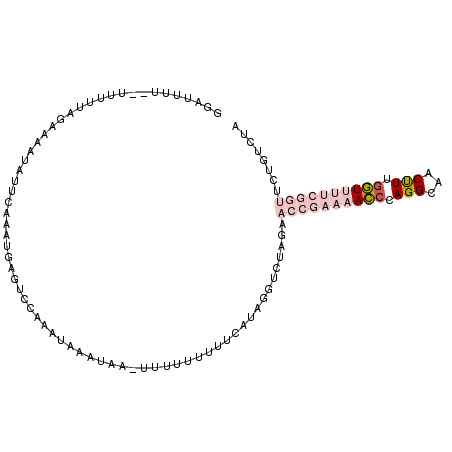
>2R_DroMel_CAF1 9031068 106 - 20766785 GGAUUUUAUUUUUUAGAAUAUAUUCAAAUGAGUCCAAAUAAAUAAAUUUUUUUUUCAUAGGUCUAGAACCGAAAACCCAGGUAACUUUGGUUUUCGGUUCUGUCUA ((((((.....(((.(((....)))))).))))))......................((((..((((((((((((((.(((...))).)))))))))))))))))) ( -26.50) >DroPse_CAF1 7636 84 - 1 AAAAUUUU-UUGU----UAAUGUUAAAAGAACUCCAAAGAAAG-----UGUGUUUCAUUGAUGUGAAACCGAAAAACCAGGCACCCUUGUU------------CUC ....((((-(((.----....(((.....)))..)))))))..-----((.(((((((....)))))))))...(((.(((...))).)))------------... ( -10.50) >DroSec_CAF1 7394 102 - 1 GGAUUUU--UUUUUAGAAAAUAUUCAAAUGAGUCCAAAUAAAUAA-UUUU-UUUUCAUAGGUCUAGAACCGAAAACACAGGUAACUUUGGUUUUCGGUUCUGUCUA ((((((.--..(((.(((....)))))).))))))..........-....-......((((..(((((((((((((.((((....))))))))))))))))))))) ( -26.00) >DroSim_CAF1 6744 102 - 1 GGAUUUU---UUUUAGAAAAUAUUCAAAUGAGUCCAAAUAAAUAA-UUUUUUUUUCAUAGGUCUAGAACCGAAAACCCAGGUAACUUUGGUUUUCGGUCCUGUCUA ((((((.---.(((.(((....)))))).))))))..........-...........((((..(((.((((((((((.(((...))).)))))))))).))))))) ( -23.40) >DroYak_CAF1 7380 99 - 1 UGUUUU---UUUUUAGAAAAUAUUCCAAUGAGUCCAGAUAGAUAA-UU---UUUUCAUAGGUAUAGAACCGAAAACCCAGGCAACUUUGGUUUUCGGUUCUGUCUA ((((((---(.....)))))))..((.(((((...((((.....)-))---).))))).)).(((((((((((((((.((....))..)))))))))))))))... ( -29.60) >DroPer_CAF1 4396 84 - 1 AAAAUUUU-UUGU----UAAUGUUAAAAGAACUCCAAAGAAAG-----UGUGUUUCAUUGAUGUGAAACCGAAAAACCAGGCACCCUUGUU------------CUC ....((((-(((.----....(((.....)))..)))))))..-----((.(((((((....)))))))))...(((.(((...))).)))------------... ( -10.50) >consensus GGAUUUU__UUUUUAGAAAAUAUUCAAAUGAGUCCAAAUAAAUAA_UUUUUUUUUCAUAGGUCUAGAACCGAAAACCCAGGCAACUUUGGUUUUCGGUUCUGUCUA ...................................................................((((((((((.(((...))).))))))))))........ ( -7.45 = -9.28 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:27 2006